Final Report Oct 2019
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Quick Guide to Free Geoinformatics for Disaster Management
Disaster Risk Reduction Research Group - Centre for Applied Geoscience School of Earth and Environmental Sciences University of Portsmouth, UK. Quick Guide to Free Geoinformatics for Disaster Management Mathias Leidig & Richard Teeuw April 2016 (version 2.3) Copyright statement: This work is published under the creative commons license 3 (CC BY-NC-SA 3.0). The license grants you to share, copy, distribute, transmit and adapt this document for non-commercial purposes under share alike conditions (http://creativecommons.org/licenses/by-nc-sa/3.0/). Please refer to this document and its authors when modifying. Disaster Risk Reduction Research Group - Centre for Applied Geoscience School of Earth and Environmental Sciences University of Portsmouth, UK. Contents Introduction ................................................................................................ 3 1. Free datasets ....................................................................................... 4 Table 1: Geospatial data sources ............................................................ 4 2. Free Software ................................................................................... 13 Table 2.1. Free GIS software .............................................................. 15 Table 2.2. Free image processing software ......................................... 17 Table 2.3. Free data viewer ................................................................. 20 Table 2.4. GoogleEarth related tools ................................................... 20 Mini -

AGD Landscape & Environment 6 (2) 2012. 76-92. COMPARISON of the MOST POPULAR OPEN-SOURCE GIS SOFTWARE in the FIELD of LANDS
AGD Landscape & Environment 6 (2) 2012. 76-92. COMPARISON OF THE MOST POPULAR OPEN-SOURCE GIS SOFTWARE IN THE FIELD OF LANDSCAPE ECOLOGY SZEMÁN ISTVÁN1 1University of Debrecen, Department of Landscape Protection and Environmental Geography, Debrecen, 4032, Egyetem tér 1. E-mail: [email protected] Received 2 March 2012; accepted in revised form 21 December 2012 Abstract GIS (Geographic Information System) software is a very useful tool in modern landscape ecology research. With its help data can be obtained which can - after processing - help to understand and demonstrate the processes taking place in the landscape. Since direct environmental measurements and sampling from a large area are, in many cases, difficult or even impossible, modelling with GIS tools is very important in the workflow of landscape research and landscape analysis. In this article we review the best known open source GIS systems and geographic information tools with possibilities for landscape ecology application. Furthermore we will introduce all the basic concepts that are associated with these open source software programmes. We provide a comparative analysis of the most widely used open source GIS applications, where, through a specific example, we will examine how these tools are used to produce basic landscape metric indicators. We will examine those functions of the programmes that are necessary to produce a complete thematic map, and finally we will emphasize various other important functions of the software to give adequate information for those users who choose open source code GIS tools, for financial reasons or otherwise, to complete a landscape ecology analysis. Our opinion is that this type of comparison is much more informative than those done by proprietary software, because these latter are all based on a basic data library, and therefore yield similar results (proj4, gdal/ogr, jts/geos, etc.) to their ‘paid’ competitors. -

Ilwis Manual
Ilwis manual click here to download The ILWIS User's Guide is intended for those who want to know how ILWIS is used in basic GIS and Image Processing operations. It trains the skills you. The User's Guide demonstrates the basic ILWIS concepts and the ILWIS user-interface, trains you in the analysis of spatial data and attribute data, teaches you. The ILWIS Applications Guide ( pp.) contains 25 discipline-oriented case studies. The case studies show advanced procedures to work with ILWIS and. ILWIS was released in June with a completely updated User's Guide. For the release of ILWIS however, there is no new User's Guide. Since the. In addition, ILWIS has new visualization functionality and a Visualization Reference Document. The ILWIS User's Guide is intended for those who want to. This ILWIS User's Guide has been rewritten and extended by Raymond Nijmeijer The ILWIS software is designed by Wim Koolhoven and Jelle Wind. LEARN ILWIS AND TEACH OTHERS IN ONE DAY ILWIS PRACTICAL MANUAL This Practical Manual Covers Just Onidex Geo-Spatial. Title, {ILWIS Academic user's guide}. Publication Type, Book. Year of Publication, Authors, Unit Geo Software Development. Number. We have special e-mail addresses for various ILWIS related subjects: This ILWIS User's Guide has been rewritten and extended by Raymond Nijmeijer. Introduction to ILWIS. Source: Cees van Westen. Associated Institute of the ILWIS Applications Guide Advanced procedures to work with ILWIS, providing case. Today I'll start with a new series of tutorials for an open Source GIS called “ILWIS GIS“. -

Open Source GIS Software Options for Forestry Education in Papua New Guinea
Open Journal of Ecology, 2014, 4, 234-243 Published Online March 2014 in SciRes. http://www.scirp.org/journal/oje http://dx.doi.org/10.4236/oje.2014.44022 Open Source GIS Software Options for Forestry Education in Papua New Guinea David Lopez Cornelio Department of Forestry, PNG University of Technology, PMB, Lae, Papua New Guinea Email: [email protected] Received 6 November 2014; revised 9 December 2014; accepted 25 December 2013 Copyright © 2014 by author and Scientific Research Publishing Inc. This work is licensed under the Creative Commons Attribution International License (CC BY). http://creativecommons.org/licenses/by/4.0/ Abstract Although open source softwares (OSS) for GIS and Remote Sensing are rapidly expanding and im- proving in the global context, there has been uncertainty at higher education institutions in de- veloping countries, such as the department of forestry (Dfo) at Unitech, Papua New Guinea (PNG), regarding appropriate GIS softwares and hardware to acquire and use for teaching and research purposes. The paper briefly describes the characteristics of some mature OSS and discusses their main capabilities, advantages and disadvantages. Their adoption in the Dfo curricula may be ad- vantageous in the long term, considering issues of learning curve steepness, versatibility, afforda- bility, effectiveness, and documentation available on them. Keywords Open Source Software; GIS Software; Forestry Education 1. Introduction GIS is a key technology for developing countries in domains such as environmental protection, urban manage- ment, agricultural production, deforestation mapping, public health assessment, and socioeconomic measure- ments. It is defined as a system of software components that maintain a spatially aware database, provides ana- lytical tools that enable spatial queries of the database, allows the association of locations with imported graphi- cal data, and provides graphical and tabular output. -

A Selection of Software for Processing Geospatial Data Is Presented in Table 1.1 and Table 1.2. Note That You May Need Some Time
A selection of software for processing geospatial data is presented in Table 1.1 and Table 1.2. Note that you may need some time to find the best software for a specific task or project. Terminology: Freeware: Freeware (a combination of the words "free" and "software") is software that is available for use at no cost or fee, but usually with one or more restricted usage rights. Compared the FOSS (see below) the source code is usually not published and hence the software can’t be modified and adjusted by 3rd parties. FOSS: Free and open-source software (F/OSS, FOSS) or free/libre/open-source software (FLOSS) is software that is both: 1) free software and 2) open source. It is licensed in the way that users are granted the right to use, copy, study, change, and improve its design through the availability of its source code. Free GIS software Description Terra MA2 (old SISM DEN) is a software product, a computational system, based on a Service Oriented Architecture (SOA), which provides the technological infrastructure required to develop operational systems for environmental risks monitoring and alert. TerraMA2 provides services to gather updated data through internet and to add it to the alert system database; services to manipulate/analyze new data in real time and check Terra MA2 if a risk situation exists by comparing with risk maps or a defined model; Monitoring, Analysis and Alert services to execute/edit/create new risk and alert models; services to create www.dpi.inpe.br/terrama2/ and notify alerts to system users; and other basic and advanced services. -

Open Access Geospatial Data Tuesday, Oct
Open Access Geospatial Data Tuesday, Oct. 25, 2016 Hannah Hamalainen Geospatial & Earth Sciences Librarian Geospatial Services Center Using Open Access Data to Make Maps is Easy Open Open source source Free Geospatial GIS Map Data software Let’s review some concepts Open Access FOSS Open Database OA removes price barriers Free and Open Source License (ODbL) (subscriptions, licensing Software which is freely Copyleft ("Share Alike") fees, pay-per-view fees) and licensed to use, copy, study, license agreement intended permission or use barriers and change the software in to allow users to freely (most copyright and any way, and the source share, modify, and use a licensing restrictions). code is openly shared. database Open Data Closed Source Open Science Open data is data that can Open source refers to is the movement to make be freely used, re-used and exclusively to software -- the scientific research, data and redistributed by anyone - source code for that software dissemination accessible to subject only, at most, to the is openly available, thus all levels of an inquiring requirement to attribute allowing for modification, and society, amateur or and sharealike. may be redistributed freely. professional. OGC The Open Geospatial Consortium, Inc (OGC) is an international industry consortium of over 387 companies, government agencies and universities participating in a consensus process to opengeospatial.org develop publicly available interface standards. Both proprietary and open source software can be compliant with OGC Open Standards OSGEO OSGEO (Open Source Geospatial Foundation) osgeo.org organization that supports development of open source software Free and Open Source Software (FOSS) QGIS ▧ AKA Quantum GIS ▧ The go-to free and open source program for geospatial analysis. -

Software (R+GIS+GE) 2
3 1 Software (R+GIS+GE) 2 This chapter will introduce you to five main packages that we will later on use in various exercises from 3 chapter 5 to 11: R, SAGA, GRASS, ILWIS and Google Earth (GE). All these are available as open source 4 or as freeware and no licenses are needed to use them. By combining the capabilities of the five software 5 packages we can operationalize preparation, processing and the visualization of the generated maps. In this 6 handbook, ILWIS GIS will be primarily used for basic editing and to process and prepare vector and raster 7 maps; SAGA/GRASS GIS will be used to run analysis on DEMs, but also for geostatistical interpolations; R 8 + packages will be used for various types of statistical and geostatistical analysis, but also for data processing 9 automation; Google Earth will be used for visualization and interpretation of results. 10 In all cases we will use R to control all pro- 11 cesses, so that each exercise will culminate in a sin- 12 gle R script (‘R on top’; Fig. 3.1). In subsequent sec- 13 tion, we will refer to the R + Open Source Desk- 14 top GIS combo of applications that combine geo- Statistical 15 computing graphical and statistical analysis and visualization as 16 R+GIS+GE. 17 GDAL This chapter is meant to serve as a sort of a 18 19 mini-manual that should help you to quickly obtain ground overlays, and install software, take first steps, and start doing time-series KML 20 some initial analysis. -

ILWIS 2.1 for Windows
ILWIS 2.1 for Windows The Integrated Land and Water Information System Reference Guide ILWIS Department, International Institute for Aerospace Survey & Earth Sciences Enschede, The Netherlands © ILWIS Department, ITC, October 1997 ITC The International Institute for Aerospace Survey and Earth Sciences, Enschede, is the largest institute for international higher education in the Netherlands. Its main objective is to assist developing countries in human resources development in aerospace surveys, Remote Sensing applications, the establishment of geoinformation systems and the management of geoinformation. To this end, ITC concentrates on three activities: education/training, research and advisory services. In-house expertise covers an extensive range of disciplines in the above fields. Disclaimer The International Institute for Aerospace Survey and Earth Sciences (ITC) has carefully prepared and reviewed this document, the software and the data set on CD-ROM for accuracy. However, ITC takes no responsibility or liability for incidental or consequential damages arising from the use of this document or the data on the accompanying CD-ROM and reserves the right to update, revise, or change this document or the data without notice. Proprietary Notice The information in this document is the sole property of the International Institute for Aerospace Survey and Earth Sciences (ITC) and may not be reproduced, stored in a retrieval system or transmitted in any form or by any means: electronic, photocopying or otherwise, without permission in writing from ITC. Contact addresses For general information about ILWIS, please contact: ILWIS Department, ITC PO Box 6, 7500 AA Enschede The Netherlands E-mail: [email protected] Tel. : +31-53-4 874 337 Fax : +31-53-4 874 484 Web-site: http://www.itc.nl/ilwis/ Remarks, suggestions and bug reports, should be sent to: Drs. -

ILWIS 2.2 for Windows
ILWIS 2.2 for Windows The Integrated Land and Water Information System Installation and New Functionality ILWIS Development, International Institute for Aerospace Survey and Earth Sciences Enschede, The Netherlands © ILWIS Development, ITC, October 1998 ITC The International Institute for Aerospace Survey and Earth Sciences, Enschede, is the largest institute for international higher education in the Netherlands. Its main objective is to assist developing countries in human resources development in aerospace surveys, Remote Sensing applications, and the establishment of geoinformation systems and the management of geoinformation. To this end, ITC concentrates on three activities: education/training, research and advisory services. In-house expertise covers an extensive range of disciplines in the above fields. PCI-ILWIS Nederland B.V. ITC & the PCI Geomatics Group started a joint venture for the distribution of geomatics software solutions and training to developing nations. This partnership resulted in the official establishment of "PCI-ILWIS Nederland B.V." As of now you are welcome to contact the PCI-ILWIS Nederland B.V. for your ILWIS orders. Disclaimer The International Institute for Aerospace Survey and Earth Sciences (ITC) has carefully prepared and reviewed this document, the software and the data set on CD-ROM for accuracy. However, ITC takes no responsibility or liability for incidental or consequential damages arising from the use of this document, the software or the data on the accompanying CD-ROM and reserves the right to update, revise, or change this document or the data without notice. Proprietary Notice The information in this document is the sole property of the International Institute for Aerospace Survey and Earth Sciences (ITC) and may not be reproduced, stored in a retrieved system, or transmitted in any form or by any means: electronically, photo copying or otherwise, without permission in writing from ITC. -
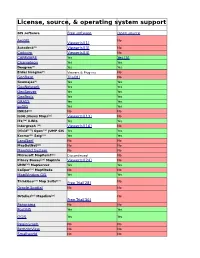
License, Source, & Operating System Support
License, source, & operating system support GIS software Free software Open source ArcGIS No Viewer(s)[1] Autodesk[2] Viewer(s)[3] No Cadcorp Viewer(s)[4] No CAPAWARE Yes Yes [5] Chameleon Yes Yes Deegree[6] Yes Yes Erdas Imagine[7] Viewers & Plug-ins No GeoBase Trial[8] No Geomajas[9] Yes Yes GeoNetwork Yes Yes GeoServer Yes Yes GeoTools Yes Yes GRASS Yes Yes gvSIG Yes Yes IDRISI[11] No No ILOG JViews Maps[12] Viewer(s)[13] No ITC[14] ILWIS Yes Yes Intergraph [15] Viewer(s)[16] No (Vivid[17]) Open[18] JUMP GIS Yes Yes Kosmo[19] Saig[20] Yes Yes LandSerf No No MapDotNet[21] No No Manifold System No No Microsoft MapPoint[22] Discontinued No Pitney Bowes[23] MapInfo Viewer(s)[24] No UMN[25] MapServer Yes Yes Caliper[26] Maptitude No No MapWindow GIS Yes Yes [27] [27] ThinkGeo Map Suite Free Trial[28] No Oracle Spatial No No Ortelius[29] Mapdiva[29] No Free Trial[30] Panorama No No PostGIS Yes Yes QGIS Yes Yes RegioGraph No No RemoteView No No Smallworld No No [31] Spatial Manager Desktop Free Trial [32] No Spatial Manager for No AutoCAD [33] Free Trial [34] SpatialFX | ObjectFX[35] No No SpatialRules | ObjectFX[35] No No SPRING Yes Yes Caliper TransCAD[26] No No TerraLib TerraView Yes Yes MicroImages[36] TNTmips Viewer(s)[37] No Caliper[38] TransModeler No No uDIG Yes Yes Zulu [39] Viewer(s) No AvisMap GIS Engine[40] No Viewer(s) GIS software Free software Open source Windows Mac OS X Linux Yes No Yes Yes No Yes Yes No S Yes No No Yes Yes Yes Java Java Java Yes No No Yes No Yes Java Java Java Java Java Java Yes Yes Yes Java Java Java Yes -

GDAL/OGR User Docs GDAL Utilities
GDAL/OGR user docs GDAL Utilities.............................................................................................................................................................1 Creating New Files.........................................................................................................................................1 General Command Line Switches..................................................................................................................2 gdalinfo.........................................................................................................................................................................3 SYNOPSIS......................................................................................................................................................3 DESCRIPTION..............................................................................................................................................3 EXAMPLE......................................................................................................................................................4 gdal_translate..............................................................................................................................................................5 SYNOPSIS......................................................................................................................................................5 DESCRIPTION..............................................................................................................................................5 -

A Feasibility Study on Applying Remotely Sensed Data for Agricultural Management in Beyza Land
International Journal of Computer Applications (0975 – 8887) Volume 99– No.8, August 2014 A Feasibility Study on Applying Remotely Sensed data for Agricultural Management in Beyza Land S. Abbas Montaseri Reza Javidan Computer Engineering Department Computer Engineering Department Beyza Branch, Islamic Azad University, Beyza Branch, Islamic Azad University, Fars, Iran Fars, Iran ABSTRACT can be summarized in three main stages: (1) Gathering Today, because of increasing demands for agricultural information using satellite imageries (2) Processing and products and existence of restrictions against resources such analyzing information to assess the significance of as lack of natural water resources for producing agricultural changeability and (3) Producing relevant outputs in forms of products using traditional methods, it is a need to use modern maps and other useful databases. techniques such as satellite remote sensing to manage natural Remote sensing today play an important role in estimating resources. The need for agriculture is to increase the crop condition and profit forecasting, domain estimates of production by optimum utilization of resources. Remote specific crops, detection of crop pests and diseases, finding sensing satellites could be a good option for water and disaster location, monitoring wild life, gathering water supply nutrient requirement and disease detection. In this paper a information, weather forecasting, management of lands, and feasibility study is carried out for using remote sensing livestock studies [2]. images for managing agricultural resources in the area of Beyza, a land in Fars province in Iran. After a study on In this paper a feasibility study is carried out for using remote availability of different imagery resources covered this area sensing images for managing agricultural resources in the area and the way of accessing such data in a good manner, ETM+ of Beyza, a land in Fars province in Iran.