Genetics Reveal the Identity and Origin of the Lionfish Invasion in The
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Documenting the Invasion of Lionfish in Florida's Waters And
Documenting the Invasion of Lionfish in Florida’s Waters and Management Efforts to Control Them Image credit: Image Bryan Fluech Scorpionfish Family Two visually identical species found in the Southeastern U.S. Red Lionfish (Pterois volitans) Devil Firefish (Pterois miles) Image credits: www.lionfishhunters.org Native Distribution Red Lionfish – Pterois volitans Devil Firefish – Pterois miles Location of Venomous Spines 13 2 3 Venomology Image Image credit: REEF Lionfish venom is a protein based neurotoxin and is contained in glandular venom tissue in grooves along either side of each spine Image credit: Dawn Witherington Lionfish Envenomation Pain is immediate; intensifies “It won’t kill you, but it’ll over 60-90 minutes; may last make you wish you were 6-12 hours dead.” Treatment involves covering the wound with hot (not scalding) water; use of over-the-counter pain relievers Victims should seek medical attention Other symptoms might include ulceration of the wound site, headaches, nausea or diarrhea Image Credit: Roxane Boonstra Lionfish Invasion First record : 1985 in Dania, FL Aquarium releases or escapes are most likely sources of the invasion Over 60,000 imported into Florida annually* Mitochondrial data show no evidence of multiple independent introductions Documenting the Invasion Image credit: USGS Lionfish Issues Image credit: Florida Sea Grant Highly Productive Year round reproduction Can spawn every 2-4 days 20,000 – 30,000 eggs / spawn Larvae dispersed by currents Sexually mature within a year Egg Mass Image -

The Invasion of Indo-Pacific Lionfish in the Bahamas: Challenges for a National Response Plan
The Invasion of Indo-Pacific Lionfish in the Bahamas: Challenges for a National Response Plan KATHLEEN SULLIVAN SEALEY1, 4, LAKESHIA ANDERSON2, DEON STEWART3, and NICOLA SMITH4, 5 1University of Miami Department of Biology, Coral Gables, Florida USA; 2 Department of Marine Resources, Nassau, Bahamas; 3 Bahamas Environment, Science and Technology Commission, Nassau, Bahamas; 4 Marine and Environmental Studies Institute, College of The Bahamas, Nassau, Bahamas; 5 Department of Zoology, University of British Columbia, Vancouver, BC, Canada ABSTRACT The invasion of the Indo-Pacific lionfish to Bahamian waters raises considerable concern due to the uncertainty of its ecological impacts and its potential threats to commercial fisheries and human safety. Lionfish have been reported throughout the archipelago and are the focus of several research and monitoring initiatives. The Bahamas has a National Invasive Species Strategy, but marine invasions require unique partnerships for small islands developing states to develop realistic management goals and actions. The Government of The Bahamas has limited funds to address major resource management issues; hence, collaboration with non-governmental agencies and tertiary education institutions is imperative. The establishment and spread of lionfish has created a novel opportunity for the formation of innovative public-private partnerships to address the ecological, economic and social impacts of biological invasions. KEY WORDS: Lionfish, invasion, reefs La Invasión en las Bahamas del Pez León del Indo-Pacifico: Un Caso de Investigación, Planificación, y Manejo La invasión del pez león del Indopacífico en las aguas de Las Bahamas ha generado una gran preocupación debido a la incertidumbre sobre su efecto ecológico y la posible afectación a la pesca comercial, el turismo y la seguridad de la población. -

A Guide to Harmful and Toxic Creatures in the Goa of Jordan
Published by the Royal Marine Conservation Society of Jordan. P. O. Box 831051, Abdel Aziz El Thaalbi St., Shmesani 11183. Amman Copyright: © The Royal Marine Conservation Society of Jordan Reproduction of this publication for educational and other non- commercial purposes is authorized without prior written approval from the copyright holder provided the source is fully acknowledged. ISBN: 978-9957-8740-1-8 Deposit Number at the National Library: 2619/6/2016 Citation: Eid, E and Al Tawaha, M. (2016). A Guide to Harmful and Toxic Creature in the Gulf of Aqaba of Jordan. The Royal Marine Conservation Society of Jordan. ISBN: 978-9957-8740-1-8. Pp 84. Material was reviewed by Dr Nidal Al Oran, International Research Center for Water, Environment and Energy\ Al Balqa’ Applied University,and Dr. Omar Attum from Indiana University Southeast at the United State of America. Cover page: Vlad61; Shutterstock Library All photographs used in this publication remain the property of the original copyright holder, and it should not be reproduced or used in other contexts without permission. 1 Content Index of Creatures Described in this Guide ......................................................... 5 Preface ................................................................................................................ 6 Part One: Introduction ......................................................................................... 8 1.1 The Gulf of Aqaba; Jordan ......................................................................... 8 1.2 Aqaba; -

Fish Biodiversity Changes in the Central Mediterranean
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Digital.CSIC Azzurro E.1*, Soto S.2, Garofalo G.3, Maynou F.2 Fistularia commersonii in the Mediterranean Sea: Invasion history and distribution modeling based on presence-only records 1ISPRA, National Institute for Environmental Protection and Research, Piazzale dei Marmi 2, 57128 Livorno, Italy 2Institut de Ciències del Mar (CSIC) Passeig Marítim de la Barceloneta, 37-49, E-08003 Barcelona, Spain 3IAMC-CNR, Institute for Coastal Marine Environment, Via L. Vaccara 61, 91026 Mazara del Vallo, Italy Running title: Fistularia commersonii in the Mediterranean Sea 1 Author to whom correspondence should be addressed. email: [email protected] Abstract The bluespotted cornetfish (Fistularia commersonii) (Osteichtyes, Fistulariidae) is considered to be one of the most invasive species of the Mediterranean Sea and Europe but only scattered information exists on its distribution and abundance. Here we collated the available species records, following its first detection in the Mediterranean Sea, in January 2000, until October 2011. A total of 191 observations were used to reconstruct the invasion sequence, to provide estimates of the rate of spread and to construct an environmental suitability model based on six biophysical variables and the maximum entropy approach (MaxEnt). The results showed that colonization of the Mediterranean Sea proceeded in parallel along the southern and northern rim of the Basin at speeds that reached 1000- 1500 km · yr-1 with a clear decrease in the rate of spread at the Sicily Strait. The most important explanatory variables for describing the distribution of F. -
Field Identification Guide to the Living Marine Resources in Kenya
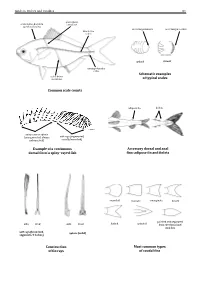
Guide to Orders and Families 81 lateral line scales above scales before dorsal fin outer margin smooth outer margin toothed (predorsal scales) lateral–line 114 scales cycloid ctenoidِّ scales circumpeduncular Schematic examples lateral line of typical scales scales below Common scale counts adipose fin finlets soft rays (segmented, spinyunbranched) rays or spines usually branched) (unsegmented, always Example of a continuous Accessory dorsal and anal dorsal fin of a spiny–rayed fish fins: adipose fin and finlets rounded truncate emarginate lunate side front side front from the dorsal and pointed and separated forked pointed soft rays (branched, spines (solid) segments, 2 halves) anal fins Construction Most common types of fin rays of caudal fins 82 Bony Fishes GUIDE TO ORDERS AND FAMILIES Order ELOPIFORMES – Tarpons and allies Fin spines absent; a single dorsal fin located above middle of body; pelvic fins in abdominal position; lateral line present; 23–25 branchiostegal rays; upper jaw extending past eye; tip of snout not overhanging mouth; colour silvery. ELOPIDAE Page 121 very small scales Ladyfishes To 90 cm. Coastal marine waters and estuaries; pelagic. A single species included in the Guide to Species.underside of head large mouth gular plate MEGALOPIDAE Page 121 last ray long Tarpons large scales To 55 cm. Coastal marine waters and estuaries; pelagic. A single species included in the Guide to Species.underside of head gular plate Order ALBULIFORMES – Bonefishes Fin spines absent; a single dorsal fin located above middle of body; pelvic fins in abdominal position; lateral line present; 6–16 branchiostegal rays; upper jaw not extending as far as front of eye; tip of snout overhanging mouth; colour silvery. -

Seamap Environmental and Biological Atlas of the Gulf of Mexico, 2017
environmental and biological atlas of the gulf of mexico 2017 gulf states marine fisheries commission number 284 february 2019 seamap SEAMAP ENVIRONMENTAL AND BIOLOGICAL ATLAS OF THE GULF OF MEXICO, 2017 Edited by Jeffrey K. Rester Gulf States Marine Fisheries Commission Manuscript Design and Layout Ashley P. Lott Gulf States Marine Fisheries Commission GULF STATES MARINE FISHERIES COMMISSION FEBRUARY 2019 NUMBER 284 This project was supported in part by the National Oceanic and Atmospheric Administration, National Marine Fisheries Service, under State/Federal Project Number NA16NMFS4350111. GULF STATES MARINE FISHERIES COMMISSION COMMISSIONERS ALABAMA Chris Blankenship John Roussel Alabama Department of Conservation 1221 Plains Port Hudson Road and Natural Resources Zachary, LA 70791 64 North Union Street Montgomery, AL 36130-1901 MISSISSIPPI Joe Spraggins, Executive Director Representative Steve McMillan Mississippi Department of Marine Resources P.O. Box 337 1141 Bayview Avenue Bay Minette, AL 36507 Biloxi, MS 39530 Chris Nelson TBA Bon Secour Fisheries, Inc. P.O. Box 60 Joe Gill, Jr. Bon Secour, AL 36511 Joe Gill Consulting, LLC 910 Desoto Street FLORIDA Ocean Springs, MS 39566-0535 Eric Sutton FL Fish and Wildlife Conservation Commission TEXAS 620 South Meridian Street Carter Smith, Executive Director Tallahassee, FL 32399-1600 Texas Parks and Wildlife Department 4200 Smith School Road Representative Jay Trumbull Austin, TX 78744 State of Florida House of Representatives 402 South Monroe Street Troy B. Williamson, II Tallahassee, FL 32399 P.O. Box 967 Corpus Christi, TX 78403 TBA Representative Wayne Faircloth LOUISIANA Texas House of Representatives Jack Montoucet, Secretary 2121 Market Street, Suite 205 LA Department of Wildlife and Fisheries Galveston, TX 77550 P.O. -

Fish Movement in the Red Sea and Implications for Marine Protected Area Design
Fish Movement in the Red Sea and Implications for Marine Protected Area Design Thesis by Irene Antonina Salinas Akhmadeeva In Partial Fulfillment of the Requirements For the Degree of Master of Science King Abdullah University of Science and Technology Thuwal, Kingdom of Saudi Arabia April, 2021 2 EXAMINATION COMMITTEE PAGE The thesis of Irene Antonina Salinas Akhmadeeva is approved by the examination committee. Committee Chairperson: Prof. Michael L. Berumen Committee Co-Chair: Dr. Alison Green Committee Members: Dr. Darren Coker, Prof. Rusty Brainard 3 COPYRIGHT © April 2021 Irene Antonina Salinas Akhmadeeva All Rights Reserved 4 ABSTRACT Fish Movement in the Red Sea and Implications for Marine Protected Area Design Irene Antonina Salinas Akhmadeeva The Red Sea is valued for its biodiversity and the livelihoods it provides for many. It now faces overfishing, habitat degradation, and anthropogenic induced climate-change. Marine Protected Areas (MPAs) became a powerful management tool to protect vulnerable species and ecosystems, re-establish their balance, and enhance marine populations. For this, they need to be well designed and managed. There are 15 designated MPAs in the Red Sea but their level of enforcement is unclear. To design an MPA it is necessary to know if it will protect species of interest by considering their movement needs. In this thesis I aim at understanding fish movement in the Red Sea, specifically home range (HR) to inform MPA size designation. With not much empirical data available on HR for Red Sea fish, I used a Machine Learning (ML) classification model, trained with empirical literature HR measurements with Maximum Total Length (L Max), Aspect Ratio (AR) of the caudal fin, and Trophic Level as predictor variables. -

Pterois Miles (Devil Firefish Or Common Lionfish)
UWI The Online Guide to the Animals of Trinidad and Tobago Diversity Pterois miles (Devil Firefish or Common Lionfish) Family: Scorpaenidae (Scorpionfish) Order: Scorpaeniformes (Mail-cheeked Fish) Class: Actinopterygii (Ray-finned fishes) Fig. 1. Devil firefish, Pterois miles. [http://pre.apaaweb.com/jordania_2009.html, downloaded 8 October 2016] TRAITS. The devil firefish or common lionfish Pterois miles has maroon and white coloured stripes throughout its entire body that allows them to blend into some habitats; it has elaborate dorsal, pelvic and pectoral fins and a pair of tentacles above its eyes (Fig. 1). It has cycloid (rounded) scales (Belmore, 2013), and the head is large, up to one third its body length. The firefish has 10 dorsal and 6 anal fin spines, all with venom that is produced from the enclosing skin (Schofield et al., 2012). The caudal (tail) rays are not enclosed by venom-producing skin. Males and females look exactly alike except during times of mating (Morris et al., 2009). Adults typically grow up to 30cm in length, but can reach 43cm and weigh 1.1kg (Belmore, 2013). DISTRIBUTION. The native geographical area of Pterois miles is mainly in the Indian Ocean (Fig. 2). More specifically it is found from the Indochina Peninsula to India and the Red Sea (Belmore, 2013). However, the devil firefish has been introduced and invaded other far-reaching regions of the world such as the east coast of the USA and Caribbean waters (Morris, 2009), and the Red Sea, probably migrating through the Suez Canal (Schofield et al., 2012). This species invaded Tobago’s waters a few years ago, at the tail end of the Caribbean, demonstrating its continued migration. -

Houssein ELBARAASI* and Osama ELSALINI
ACTA ICHTHYOLOGICA ET PISCATORIA (2009) 39 (1): 63–66 DOI: 10.3750/AIP2009.39.1.13 A RECORD OF BLUESPOTTED CORNETFISH, FISTULARIA COMMERSONII (ACTINOPTERYGII: SYNGNATHIFORMES: FISTULARIIDAE), OFF THE COAST OF BENGHAZI, LIBYA (SOUTHERN MEDITERRANEAN ) Houssein ELBARAASI * and Osama ELSALINI Department of Zoology, Faculty of Science, Garyounis University, Benghazi, Libya Elbaraasi H., ElSalini O. 2009. A record of bluespotted cornetfish , Fistularia commersonii (Actinopterygii : Syngnathiformes: Fistulariidae), off the coast of Benghazi, Libya (southern Mediterranean ). Acta Ichthyol. Piscat. 39 (1): 63–66. Abstract. A record of bluespotted cornetfish Fistularia commersonii Rüppell, 1835, (Fistulariidae) from the depth of 30 m, caught off the coast of Benghazi, Libya (Southern Mediterranean), is reported herewith. Keywords: bluespotted cornetfish, Fistularia commersonii , first record, Mediterranean, Libya The opening of Suez Canal in 1869 connected the Red the Algerian coasts, in Skikda Bay (eastern Algeria) and Sea to the Mediterranean and allowed the introduction of offshore of Bou Ismail Bay (central Algeria) at the numerous Indo-Pacific species into the Mediterranean Mediterranean (Kara and Oudjane 2008). (Golani 1998a, 1998b, Kasapidis et al. 2007), including In this short note, a record of the bluespotted cornetfish, bluespotted cornetfish, Fistularia commersonii Rüppell, 1835 F. commersonii , in the Libyan waters is reported. (cf. Golani 2000, Karachle et al. 2004, Golani et al. 2007). The specimen of F. commersonii was caught in The bluespotted cornetfish F. commersonii is distrib - November 2007 by a commercial bottom trawl, 5 nautical uted in tropical and subtropical seas, common among miles off the coast of Benghazi, Southern Mediterranean, reefs, shallow sandy bottoms, and on sea grass beds Libya, (lat 32°06'N, long 20°03'E, Fig. -

Competitive Interactions for Shelter Between Invasive Pacific Red Lionfish and Native Nassau Grouper
Environ Biol Fish DOI 10.1007/s10641-014-0236-9 Competitive interactions for shelter between invasive Pacific red lionfish and native Nassau grouper Wendel W.Raymond & Mark A. Albins & Timothy J. Pusack Received: 12 September 2013 /Accepted: 14 January 2014 # Springer Science+Business Media Dordrecht 2014 Abstract The invasive Pacific red lionfish (Pterois positioned closer to and used limited shelter more when volitans) poses a threat to western Atlantic and Carib- paired with similarly sized lionfish and (2) grouper bean coral reef systems. Lionfish are small-bodied pred- moved much further away from shelter when paired ators that can reduce the abundance and diversity of with smaller lionfish. We also found that neither large native fishes via predation. Additionally, native preda- lionfish nor large Nassau grouper preyed upon smaller tors or competitors appear to have a negligible effect on individuals of the opposite species suggesting that Nas- similarly sized lionfish. Nassau grouper (Epinephelus sau grouper do not recognize small lionfish as prey. This striatus) are a regionally endangered, large predator study highlights how invasive lionfish may affect native found throughout lionfish’s invasive range. Because Nassau grouper, and suggests that competition for shel- lionfish and Nassau grouper occupy similar habitats ter between these two species may be size dependent. and use similar resources, there is potential for compe- tition between these two species. Using large, outdoor Keywords Avoidance . Bahamas . Refuge . Shelter in-ground tanks, we investigated how lionfish and Nas- dominance . Size dependence sau grouper affect each other’s behavior by comparing their distance from and use of shelter when in isolation versus when both species were in the presence of each Introduction other with limited shelter. -

Combined Analysis of Four Mitochondrial Regions Allowed The
Journal of the Marine Biological Association of the United Kingdom, page 1 of 5. # Marine Biological Association of the United Kingdom, 2010 doi:10.1017/S0025315410001451 Combined analysis of four mitochondrial regions allowed the detection of several matrilineal lineages of the lessepsian fish Fistularia commersonii in the Mediterranean Sea daria sanna1, paolo merella2, tiziana lai1, sarra farjallah3, paolo francalacci1, marco curini-galletti1, antonio pais4 and marco casu1 1Dipartimento di Zoologia e Genetica Evoluzionistica, Universita` di Sassari, Via F. Muroni 25, 07100 Sassari, Italy, 2Sezione di Parassitologia e Malattie Parassitarie, Dipartimento di Biologia Animale, Universita` di Sassari, Via Vienna 2, 07100 Sassari, Italy, 3Unite´ de Recherche: Ge´ne´tique, Biodiversite´ et Valorisation des Bioressources UR/09-30, Institut Supe´rieur de Biotechnologie de Monastir, Monastir 5000, Tunisia, 4Sezione di Acquacoltura e Gestione delle Risorse Acquatiche, Dipartimento di Scienze Zootecniche, Universita` di Sassari, Via E. De Nicola 9, 07100 Sassari, Italy The bluespotted cornetfish (Fistularia commersonii) is an Indo-Pacific species that in the last ten years colonized a large part of the Mediterranean basin. The aim of this study was to sequence some portions of the mitochondrial DNA (D-loop II, 16S, 12S and Cyt b) of this fish from different localities of the Mediterranean Sea, in order to evaluate the level of its genetic varia- bility in this area. The genetic analysis performed on specimens from seven localities of Sardinia, Tunisia and Libya revealed the presence of at least five mitochondrial lineages. The results obtained, compared with previous studies, indicate that the use of a sufficient number of mitochondrial regions may allow a more accurate estimate of genetic variability in lessepsian invasions. -

Pterois Miles) Ecological Risk Screening Summary
U.S. Fish and Wildlife Service Devil Firefish (Pterois miles) Ecological Risk Screening Summary Web Version—07/28/2014 Photo: © J.E. Randall from EOL (2014). 1 Native Range, and Status in the United States Native Range From Eschmeyer (1986): “Indian Ocean: Red Sea south to Port Alfred, South Africa and east to Sumatra, Indonesia (Fricke 1999).” Pterois miles Ecological Risk Screening Summary U.S. Fish and Wildlife Service – Web Version – 7/28/2014 Status in the United States From Schofield et al. (2014): “Atlantic Coast of USA: Lionfishes have been established from Miami to North Carolina since 2002. They established in the Florida Keys in 2009. Although present in Atlantic waters north of North Carolina, they are not likely to survive cold winter temperatures.” “Gulf of Mexico: Other than the anomalous Treasure Island specimen (see Schofield 2010), the first confirmed specimens of lionfish taken from the Gulf of Mexico were in December 2009. Sightings of lionfishes are becoming common in the northern Gulf of Mexico, especially associated with [artificial] reefs (including oil/gas platforms).” “Greater Antilles: Lionfishes are established off all islands in the Greater Antilles (Cuba [2007], Jamaica [2008], Hispañola [Haiti and the Dominican Republic; 2008] and Puerto Rico [2009]).” “Lesser Antilles: Lionfish presence has been confirmed throughout the leeward and windward islands. For more details, see Schofield (2010).” Means of Introductions in the United States From Schofield et al. (2014): “The most probable explanation for the arrival of lionfishes in the Atlantic Ocean is via the aquarium trade (Whitfield et al. 2002; Semmens et al. 2004). No one will ever know with certainty how lionfishes gained entry to the coastal waters of the U.S.; however, as they are common aquarium fishes, it is possible they were released pets.