High Prevalence, Coinfection Rate, and Genetic Diversity of Retroviruses in Wild Red Colobus Monkeys
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Hunting, Law Enforcement, and African Primate Conservation
Research Note Hunting, Law Enforcement, and African Primate Conservation PAUL K. N’GORAN,∗†‡§ CHRISTOPHE BOESCH,∗†‡ ROGER MUNDRY,∗ ELIEZER K. N’GORAN,∗∗ ILKA HERBINGER,‡ FABRICE A. YAPI,†† AND HJALMAR S. KUHL¨ ∗‡‡ ∗Department of Primatology, Max Planck Institute for Evolutionary Anthropology, Deutscher Platz 6, 04103 Leipzig, Germany †Centre Suisse de Recherches Scientifiques en Cote-d’Ivoire,ˆ 01 BP 1303 Abidjan 01, Coteˆ d’Ivoire ‡Wild Chimpanzee Foundation s/c CSRS 01 BP 1303 Abidjan 01, Coteˆ d’Ivoire §UFR Sciences de la Nature, Universite´ d’Abobo-Adjame,´ 02 BP 801 Abidjan 02, Coteˆ d’Ivoire ∗∗UFR Biosciences, Universite´ de Cocody, 01 BP V34 Abidjan 01, Coteˆ d’Ivoire ††Office Ivoirien des Parcs et Reserves,´ Direction de Zone Sud-Ouest, BP 1342 Soubre,´ Coteˆ d’Ivoire Abstract: Primates are regularly hunted for bushmeat in tropical forests, and systematic ecological monitor- ing can help determine the effect hunting has on these and other hunted species. Monitoring can also be used to inform law enforcement and managers of where hunting is concentrated. We evaluated the effects of law enforcement informed by monitoring data on density and spatial distribution of 8 monkey species in Ta¨ıNa- tional Park, Coteˆ d’Ivoire. We conducted intensive surveys of monkeys and looked for signs of human activity throughout the park. We also gathered information on the activities of law-enforcement personnel related to hunting and evaluated the relative effects of hunting, forest cover and proximity to rivers, and conservation effort on primate distribution and density. The effects of hunting on monkeys varied among species. Red colobus monkeys (Procolobus badius) were most affected and Campbell’s monkeys (Cercopithecus campbelli) were least affected by hunting. -
Online Appendix for “The Impact of the “World's 25 Most Endangered
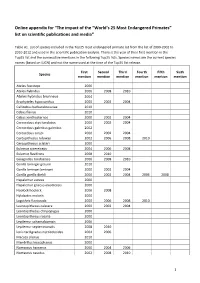
Online appendix for “The impact of the “World’s 25 Most Endangered Primates” list on scientific publications and media” Table A1. List of species included in the Top25 most endangered primate list from the list of 2000-2002 to 2010-2012 and used in the scientific publication analysis. There is the year of their first mention in the Top25 list and the consecutive mentions in the following Top25 lists. Species names are the current species names (based on IUCN) and not the name used at the time of the Top25 list release. First Second Third Fourth Fifth Sixth Species mention mention mention mention mention mention Ateles fusciceps 2006 Ateles hybridus 2006 2008 2010 Ateles hybridus brunneus 2004 Brachyteles hypoxanthus 2000 2002 2004 Callicebus barbarabrownae 2010 Cebus flavius 2010 Cebus xanthosternos 2000 2002 2004 Cercocebus atys lunulatus 2000 2002 2004 Cercocebus galeritus galeritus 2002 Cercocebus sanjei 2000 2002 2004 Cercopithecus roloway 2002 2006 2008 2010 Cercopithecus sclateri 2000 Eulemur cinereiceps 2004 2006 2008 Eulemur flavifrons 2008 2010 Galagoides rondoensis 2006 2008 2010 Gorilla beringei graueri 2010 Gorilla beringei beringei 2000 2002 2004 Gorilla gorilla diehli 2000 2002 2004 2006 2008 Hapalemur aureus 2000 Hapalemur griseus alaotrensis 2000 Hoolock hoolock 2006 2008 Hylobates moloch 2000 Lagothrix flavicauda 2000 2006 2008 2010 Leontopithecus caissara 2000 2002 2004 Leontopithecus chrysopygus 2000 Leontopithecus rosalia 2000 Lepilemur sahamalazensis 2006 Lepilemur septentrionalis 2008 2010 Loris tardigradus nycticeboides -

Abstract Book
EAZA NUTRITION GROUP & ZOOLOGICKÁ ZAHRADA LIBEREC ABSTRACT BOOK Editors Liberec Zoological Garden Marcus Clauss Lidové sady 425/1 Anouk Fens 460 01 Liberec 1 Joeke Nijboer Czech Republic Foreword Dear friends and colleagues, Time flies! After the first European Zoo Nutrition Conference in Rotterdam, in 1999, we are happy to present you the 9th European Zoo Nutrition Conference in Liberec, the Czech Republic. The European Nutrition Group (ENG) has proven to be an active group within EAZA. Certainly, it means that the animals in the European zoos will benefit from the improved diets as a result of presentations, posters, workshops and discussions held at the conferences and by the other activities organised by the ENG. One of the promoters of the ENG, Andrea Fidgett, resigned after chairing the group for more than 10 years, as she accepted a position in the USA. Ollie Szyszka, nutritionist in Marwell Zoo, will be the new chair and intends to stimulate and promote zoo animal nutrition within the ENG and European zoo community. The organising committee is pleased to present you a wide range of talks and posters, varying from sustainable palm oil, rhinoceros feeding, contraception by feeding, dietary drift, fertility in birds, milk composition, insects as feed for zoo animals to pest control in zoos. Prior to the conference, the EAZA Academy in cooperation with ENG, organises a workshop on January 26th titled: pellet formulation. After the success of the practical workstations during the last conference in Arnhem Zoo, we will have several workstation again during the zoo visit on Saturday, where information will be presented on specific zoo animal nutrition items during. -

World's Most Endangered Primates
Primates in Peril The World’s 25 Most Endangered Primates 2016–2018 Edited by Christoph Schwitzer, Russell A. Mittermeier, Anthony B. Rylands, Federica Chiozza, Elizabeth A. Williamson, Elizabeth J. Macfie, Janette Wallis and Alison Cotton Illustrations by Stephen D. Nash IUCN SSC Primate Specialist Group (PSG) International Primatological Society (IPS) Conservation International (CI) Bristol Zoological Society (BZS) Published by: IUCN SSC Primate Specialist Group (PSG), International Primatological Society (IPS), Conservation International (CI), Bristol Zoological Society (BZS) Copyright: ©2017 Conservation International All rights reserved. No part of this report may be reproduced in any form or by any means without permission in writing from the publisher. Inquiries to the publisher should be directed to the following address: Russell A. Mittermeier, Chair, IUCN SSC Primate Specialist Group, Conservation International, 2011 Crystal Drive, Suite 500, Arlington, VA 22202, USA. Citation (report): Schwitzer, C., Mittermeier, R.A., Rylands, A.B., Chiozza, F., Williamson, E.A., Macfie, E.J., Wallis, J. and Cotton, A. (eds.). 2017. Primates in Peril: The World’s 25 Most Endangered Primates 2016–2018. IUCN SSC Primate Specialist Group (PSG), International Primatological Society (IPS), Conservation International (CI), and Bristol Zoological Society, Arlington, VA. 99 pp. Citation (species): Salmona, J., Patel, E.R., Chikhi, L. and Banks, M.A. 2017. Propithecus perrieri (Lavauden, 1931). In: C. Schwitzer, R.A. Mittermeier, A.B. Rylands, F. Chiozza, E.A. Williamson, E.J. Macfie, J. Wallis and A. Cotton (eds.), Primates in Peril: The World’s 25 Most Endangered Primates 2016–2018, pp. 40-43. IUCN SSC Primate Specialist Group (PSG), International Primatological Society (IPS), Conservation International (CI), and Bristol Zoological Society, Arlington, VA. -

Gastrointestinal Parasites of the Colobus Monkeys of Uganda
J. Parasitol., 91(3), 2005, pp. 569±573 q American Society of Parasitologists 2005 GASTROINTESTINAL PARASITES OF THE COLOBUS MONKEYS OF UGANDA Thomas R. Gillespie*², Ellis C. Greiner³, and Colin A. Chapman²§ Department of Zoology, University of Florida, Gainesville, Florida 32611. e-mail: [email protected] ABSTRACT: From August 1997 to July 2003, we collected 2,103 fecal samples from free-ranging individuals of the 3 colobus monkey species of UgandaÐthe endangered red colobus (Piliocolobus tephrosceles), the eastern black-and-white colobus (Co- lobus guereza), and the Angolan black-and-white colobus (C. angolensis)Ðto identify and determine the prevalence of gastro- intestinal parasites. Helminth eggs, larvae, and protozoan cysts were isolated by sodium nitrate ¯otation and fecal sedimentation. Coprocultures facilitated identi®cation of helminths. Seven nematodes (Strongyloides fulleborni, S. stercoralis, Oesophagostomum sp., an unidenti®ed strongyle, Trichuris sp., Ascaris sp., and Colobenterobius sp.), 1 cestode (Bertiella sp.), 1 trematode (Dicro- coeliidae), and 3 protozoans (Entamoeba coli, E. histolytica, and Giardia lamblia) were detected. Seasonal patterns of infection were not apparent for any parasite species infecting colobus monkeys. Prevalence of S. fulleborni was higher in adult male compared to adult female red colobus, but prevalence did not differ for any other shared parasite species between age and sex classes. Colobinae is a large subfamily of leaf-eating, Old World 19 from Angolan black-and-white colobus. Red colobus are sexually monkeys represented in Africa by species of 3 genera, Colobus, dimorphic, with males averaging 10.5 kg and females 7.0 kg (Oates et al., 1994); they display a multimale±multifemale social structure and Procolobus, and Piliocolobus (Grubb et al., 2002). -

Primate Conservation 2006 (20): 1–28
Contents General Primates in Peril: The World’s 25 Most Endangered Primates, 2004–2006 ..................................................................................1 Russell A. Mittermeier, Cláudio Valladares-Pádua, Anthony B. Rylands, Ardith A. Eudey, Thomas M. Butynski, Jörg U. Ganzhorn, Rebecca Kormos, John M. Aguiar and Sally Walker Neotropical Region On a New Species of Titi Monkey, Genus Callicebus Thomas (Primates, Pitheciidae), from Western Bolivia with Preliminary Notes on Distribution and Abundance ...............................................................................................................29 Robert. B. Wallace, Humberto Gómez, Annika Felton and Adam M. Felton Identifi cation, Behavioral Observations, and Notes on the Distribution of the Titi Monkeys Callicebus modestus Lönnberg, 1939 and Callicebus olallae Lönnberg, 1939 ..............................................................................41 Adam Felton, Annika M. Felton, Robert B. Wallace and Humberto Gómez A Survey of Primate Populations in Northeastern Venezuelan Guayana .....................................................................................47 Bernardo Urbani A History of Long-term Research and Conservation of Northern Muriquis (Brachyteles hypoxanthus) at the Estação Biológica de Caratinga/RPPN-FMA .......................................................................................................................53 Karen B. Strier and Jean Philippe Boubli Africa English Common Names for Subspecies and Species of African Primates -

Primate Occurrence Across a Human- Impacted Landscape In
Primate occurrence across a human- impacted landscape in Guinea-Bissau and neighbouring regions in West Africa: using a systematic literature review to highlight the next conservation steps Elena Bersacola1,2, Joana Bessa1,3, Amélia Frazão-Moreira1,4, Dora Biro3, Cláudia Sousa1,4,† and Kimberley Jane Hockings1,4,5 1 Centre for Research in Anthropology (CRIA/NOVA FCSH), Lisbon, Portugal 2 Anthropological Centre for Conservation, the Environment and Development (ACCEND), Department of Humanities and Social Sciences, Oxford Brookes University, Oxford, United Kingdom 3 Department of Zoology, University of Oxford, Oxford, United Kingdom 4 Department of Anthropology, Faculty of Social Sciences and Humanities, Universidade NOVA de Lisboa, Lisbon, Portugal 5 Centre for Ecology and Conservation, College of Life and Environmental Sciences, University of Exeter, Cornwall, United Kingdom † Deceased. ABSTRACT Background. West African landscapes are largely characterised by complex agroforest mosaics. Although the West African forests are considered a nonhuman primate hotspot, knowledge on the distribution of many species is often lacking and out- of-date. Considering the fast-changing nature of the landscapes in this region, up- to-date information on primate occurrence is urgently needed, particularly of taxa such as colobines, which may be more sensitive to habitat modification than others. Understanding wildlife occurrence and mechanisms of persistence in these human- dominated landscapes is fundamental for developing effective conservation strategies. Submitted 2 March 2018 Accepted 6 May 2018 Methods. In this paper, we aim to review current knowledge on the distribution of Published 23 May 2018 three threatened primates in Guinea-Bissau and neighbouring regions, highlighting Corresponding author research gaps and identifying priority research and conservation action. -

Bioko Red Colobus Piliocolobus Pennantii Pennantii (Waterhouse, 1838) Bioko Island, Equatorial Guinea (2004, 2006, 2010, 2012)
Bioko Red Colobus Piliocolobus pennantii pennantii (Waterhouse, 1838) Bioko Island, Equatorial Guinea (2004, 2006, 2010, 2012) Drew T. Cronin, Gail W. Hearn & John F. Oates Bioko red colobus (Piliocolobus pennantii pennantii) (Illustration: Stephen D. Nash) Pennant’s red colobus monkey Piliocolobus pennantii is P. p. pennantii is threatened by bushmeat hunting, presently regarded by the IUCN Red List as comprising most notably since the early 1980s when a commercial three subspecies: P. pennantii pennantii of Bioko, P. p. bushmeat market appeared in the town of Malabo epieni of the Niger Delta, and P. p. bouvieri of the Congo (Butynski and Koster 1994). Following the discovery Republic. Some accounts give full species status to of offshore oil in 1996, and the subsequent expansion all three of these monkeys (Groves 2007; Oates 2011; of Equatorial Guinea’s economy, rising urban demand Groves and Ting 2013). P. p. pennantii is currently led to increased numbers of primate carcasses in the classified as Endangered (Oates and Struhsaker 2008). bushmeat market (Morra et al. 2009; Cronin 2013). In November 2007, a primate hunting ban was enacted Piliocolobus pennantii pennantii may once have occurred on Bioko, but it lacked any realistic enforcement and over most of Bioko, but it is now probably limited to an contributed to a spike in the numbers of monkeys in the area of less than 300 km² within the Gran Caldera and market. Between October 1997 and September 2010, a 510 km² range in the Southern Highlands Scientific a total of 1,754 P. p. pennantii were observed for sale Reserve (GCSH) (Cronin et al. -

Piliocolobus Semlikiensis, Semliki Red Colobus
The IUCN Red List of Threatened Species™ ISSN 2307-8235 (online) IUCN 2020: T92657343A92657454 Scope(s): Global Language: English Piliocolobus semlikiensis, Semliki Red Colobus Assessment by: Maisels, F. & Ting, N. View on www.iucnredlist.org Citation: Maisels, F. & Ting, N. 2020. Piliocolobus semlikiensis. The IUCN Red List of Threatened Species 2020: e.T92657343A92657454. https://dx.doi.org/10.2305/IUCN.UK.2020- 1.RLTS.T92657343A92657454.en Copyright: © 2020 International Union for Conservation of Nature and Natural Resources Reproduction of this publication for educational or other non-commercial purposes is authorized without prior written permission from the copyright holder provided the source is fully acknowledged. Reproduction of this publication for resale, reposting or other commercial purposes is prohibited without prior written permission from the copyright holder. For further details see Terms of Use. The IUCN Red List of Threatened Species™ is produced and managed by the IUCN Global Species Programme, the IUCN Species Survival Commission (SSC) and The IUCN Red List Partnership. The IUCN Red List Partners are: Arizona State University; BirdLife International; Botanic Gardens Conservation International; Conservation International; NatureServe; Royal Botanic Gardens, Kew; Sapienza University of Rome; Texas A&M University; and Zoological Society of London. If you see any errors or have any questions or suggestions on what is shown in this document, please provide us with feedback so that we can correct or extend the information provided. THE IUCN RED LIST OF THREATENED SPECIES™ Taxonomy Kingdom Phylum Class Order Family Animalia Chordata Mammalia Primates Cercopithecidae Scientific Name: Piliocolobus semlikiensis (Colyn, 1991) Synonym(s): • Colobus ellioti Dollman, 1909 • Colobus variabilis Lorenz von Liburnau, 1914 • Colobus badius ssp. -

East Africa Highlights: Kenya & Tanzania I 2019
Field Guides Tour Report East Africa Highlights: Kenya & Tanzania I 2019 Mar 1, 2019 to Mar 21, 2019 Terry Stevenson For our tour description, itinerary, past triplists, dates, fees, and more, please VISIT OUR TOUR PAGE. This Gray Crowned-Crane perched atop a tree, giving us a wonderful view of this impressive bird. Photo by participant Ken Havard. Our March 2019 East Africa Highlights tour was one of total contrasts, with Tanzania being lush, wet, and green, while Kenya was dry to the extreme. All this made for some unusual and interesting bird and mammals sightings, as we traveled from Ngorongoro Crater and the Serengeti, to Tarangire, Lake Nakuru, Kakamega Forest, Baringo and Mt. Kenya. Beginning in Nairobi, we made an afternoon visit to the nearby national park where our first lions were lying right beside the road. Giraffe, Burchell's Zebra, African Buffalo, Warthog, and a variety of the more common gazelles were all part of a truly African scene as we meandered across the plains and through the acacia scrub. The birding also got off to a great start with everyone enjoying Common Ostrich (our first of many), Helmeted Guineafowl, White-bellied Go-away-bird, Black Crake, Long-toed Lapwing, Saddle-billed Stork, African Darter, Hamerkop, Goliath Heron, Black- winged Kite, Speckled Mousebird, African Gray Hornbill, Long-tailed Shrike, Northern Pied-Babbler, Red-billed Oxpecker, and Variable Sunbird. We then flew to Kilimanjaro airport and began our journey west to Ngorongoro and the Serengeti. Highlights began with a walk in the forest above Gibb's Farm where we found Schalow's Turaco, African Emerald Cuckoo, Crowned Eagle, Cinnamon-chested Bee-eater, Scaly-throated Honeyguide, African Broadbill, Black-throated Wattle-eye, White-tailed Blue-Flycatcher, Brown-headed Apalis, Gray-capped Warbler, and Thick-billed Seedeater. -

Paternity Data and Relative Testes Size As Measures of Level of Sperm Competition in the Cercopithecoidea
Received: 23 August 2018 | Revised: 10 October 2018 | Accepted: 4 November 2018 DOI: 10.1002/ajp.22937 RESEARCH ARTICLE Paternity data and relative testes size as measures of level of sperm competition in the Cercopithecoidea R. Robin Baker1 | Todd K. Shackelford2 1 School of Biological Sciences, University of Manchester, Manchester, UK Historically, the empirical study of the role of sperm competition in the evolution of 2 Department of Psychology, Oakland sexual traits has been problematic through an enforced reliance on indirect proxy University, Rochester, Michigan measures. Recently, however, a procedure was developed that uses paternity data to Correspondence measure sperm competition level directly in terms of males/conception (i.e., the Todd K. Shackelford, Department of Psychology, Oakland University, 112 Pryale number of males that have sperm present in a female's ampulla at conception). When Hal, Rochester, MI 48309-4401. tested on apes and humans (Hominoidea) this measure proved not only to correlate Email: [email protected] significantly with the traditionally used measure of relative testes size but also to offer a number of advantages. Here we provide a second test of the procedure, this time using paternity data for the Old World monkeys (Cercopithecoidea). We calculate sperm competition levels (males/conception) for 17 species of wild and free-ranging cercopithecoids and then analyze the data against measures of relative testes size. Calculated sperm competition levels correlate strongly with relative testes size both with and without phylogenetic control at both the species and generic levels. The signal-to-noise ratios inherent in both the past measure of relative testes size and the new measure of sperm competition level from paternity data are discussed. -

Colobus Guereza
Lauck et al. Retrovirology 2013, 10:107 http://www.retrovirology.com/content/10/1/107 RESEARCH Open Access Discovery and full genome characterization of two highly divergent simian immunodeficiency viruses infecting black-and-white colobus monkeys (Colobus guereza) in Kibale National Park, Uganda Michael Lauck1, William M Switzer2, Samuel D Sibley3, David Hyeroba4, Alex Tumukunde4, Geoffrey Weny4, Bill Taylor5, Anupama Shankar2, Nelson Ting6, Colin A Chapman4,7,8, Thomas C Friedrich1,3, Tony L Goldberg1,3,4 and David H O'Connor1,9* Abstract Background: African non-human primates (NHPs) are natural hosts for simian immunodeficiency viruses (SIV), the zoonotic transmission of which led to the emergence of HIV-1 and HIV-2. However, our understanding of SIV diversity and evolution is limited by incomplete taxonomic and geographic sampling of NHPs, particularly in East Africa. In this study, we screened blood specimens from nine black-and-white colobus monkeys (Colobus guereza occidentalis) from Kibale National Park, Uganda, for novel SIVs using a combination of serology and “unbiased” deep-sequencing, a method that does not rely on genetic similarity to previously characterized viruses. Results: We identified two novel and divergent SIVs, tentatively named SIVkcol-1 and SIVkcol-2, and assembled genomes covering the entire coding region for each virus. SIVkcol-1 and SIVkcol-2 were detected in three and four animals, respectively, but with no animals co-infected. Phylogenetic analyses showed that SIVkcol-1 and SIVkcol-2 form a lineage with SIVcol, previously discovered in black-and-white colobus from Cameroon. Although SIVkcol-1 and SIVkcol-2 were isolated from the same host population in Uganda, SIVkcol-1 is more closely related to SIVcol than to SIVkcol-2.