Electronic Supplementary Information
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Download Article PDF/Slides
Kan Lu, PharmD New Antiretrovirals for Based on a presentation at prn by Roy M. Gulick, md, mph the Treatment of HIV: Kan Lu, PharmD | Drug Development Fellow University of North Carolina School of Pharmacy Chapel Hill, North Carolina The View in 2006 Roy M. Gulick, md, mph Reprinted from The prn Notebook® | october 2006 | Dr. James F. Braun, Editor-in-Chief Director, Cornell Clinical Trials Unit | Associate Professor of Medicine, Meri D. Pozo, PhD, Managing Editor. Published in New York City by the Physicians’ Research Network, Inc.® Weill Medical College of Cornell University | New York, New York John Graham Brown, Executive Director. For further information and other articles available online, visit http://www.prn.org | All rights reserved. ©october 2006 substantial progress continues to be made in the arena of cokinetics and a long extracellular half-life of approximately 10 hours antiretroviral drug development. prn is again proud to present its annual (Zhu, 2003). During apricitabine’s development, a serious drug interac- review of the experimental agents to watch for in the coming months and tion with lamivudine (Epivir) was noted. Although the plasma years. This year’s review is based on a lecture by Dr. Roy M. Gulick, a long- concentrations of apricitabine were unaffected by coadministration of time friend of prn, and no stranger to the antiretroviral development lamivudine, the intracellular concentrations of apricitabine were reduced pipeline. by approximately sixfold. Additionally, the 50% inhibitory concentration To date, twenty-two antiretrovirals have been approved by the Food (ic50) of apricitabine against hiv with the M184V mutation was increased and Drug Administration (fda) for the treatment of hiv infection. -

Download Article PDF/Slides
New Antiretrovirals in Development: Reprinted from The PRN Notebook,™ june 2002. Dr. James F. Braun, Editor-in-Chief. Tim Horn, Executive Editor. Published in New York City by the Physicians’ Research Network, Inc.,® John Graham Brown, Executive Director. For further information and other articles The View in 2002 available online, visit http://www.PRN.org All rights reserved. © june 2002. Roy “Trip” Gulick, md, mph Associate Professor of Medicine, Weill Medical College of Cornell University Director, Cornell Clinical Trials Unit, New York, New York Summary by Tim Horn Edited by Scott Hammer, md espite the fact that 16 antiretro- tiviral activity of emtricitabine was estab- Preliminary results from two random- virals are approved for use in the lished, with total daily doses of 200 mg or ized studies—FTC-302 and FTC-303—were United States, there is an indis- more producing the greatest median viral reported by Dr. Charles van der Horst and putable need for new anti-hiv com- load suppression: 1.72-1.92 log. Based on his colleagues at the 8th croi, held in Feb- pounds that have potent and these data, a once-daily dose of 200 mg ruary 2001 in Chicago (van der Horst, durable efficacy profiles, unique re- was selected for further long-term clinical 2001). FTC-302 was a blinded comparison sistance patterns, patient-friendly dosing study. “This is what we’re looking forward of emtricitabine and lamivudine, both in schedules, and minimal toxicities. To pro- to with emtricitabine,” commented Dr. combination with stavudine (Zerit) and vide prn with a glimpse of drugs current- Gulick. -

Ep 2531027 B1
(19) TZZ ¥_Z _T (11) EP 2 531 027 B1 (12) EUROPEAN PATENT SPECIFICATION (45) Date of publication and mention (51) Int Cl.: of the grant of the patent: A61K 31/4985 (2006.01) A61K 31/52 (2006.01) 06.05.2015 Bulletin 2015/19 A61K 31/536 (2006.01) A61K 31/513 (2006.01) A61K 38/55 (2006.01) A61P 31/18 (2006.01) (21) Application number: 11737484.3 (86) International application number: (22) Date of filing: 24.01.2011 PCT/US2011/022219 (87) International publication number: WO 2011/094150 (04.08.2011 Gazette 2011/31) (54) Therapeutic combination comprising dolutegravir, abacavir and lamivudine Therapeutische Zusammensetzung enthaltend Dolutegravir, Abacavir und Lamivudine Combinaison thérapeutique comprenant du dolutégravir, de l’abacavir et de la lamivudine (84) Designated Contracting States: (74) Representative: Gladwin, Amanda Rachel AL AT BE BG CH CY CZ DE DK EE ES FI FR GB GlaxoSmithKline GR HR HU IE IS IT LI LT LU LV MC MK MT NL NO Global Patents (CN925.1) PL PT RO RS SE SI SK SM TR 980 Great West Road Designated Extension States: Brentford, Middlesex TW8 9GS (GB) BA ME (56) References cited: (30) Priority: 27.01.2010 US 298589 P WO-A1-2010/011812 WO-A2-2009/148600 US-A1- 2006 084 627 US-A1- 2006 084 627 (43) Date of publication of application: US-A1- 2008 076 738 US-A1- 2009 318 421 12.12.2012 Bulletin 2012/50 US-A1- 2009 318 421 US-B1- 6 544 961 (73) Proprietor: VIIV Healthcare Company • SONG1 et al: "The Effect of Ritonavir-Boosted Research Triangle Park, NC 27709 (US) ProteaseInhibitors on the HIV Integrase Inhibitor, S/GSK1349572,in Healthy Subjects", INTERNET , (72) Inventor: UNDERWOOD, Mark, Richard 15 September 2009 (2009-09-15), XP002697436, Research Triangle Park Retrieved from the Internet: URL:http: North Carolina 27709 (US) //www.natap.org/2009/ICCAC/ICCAC_ 52.htm [retrieved on 2013-05-21] Note: Within nine months of the publication of the mention of the grant of the European patent in the European Patent Bulletin, any person may give notice to the European Patent Office of opposition to that patent, in accordance with the Implementing Regulations. -

The Design, Synthesis and Optimization of Allosteric Hiv-1
THE DESIGN, SYNTHESIS AND OPTIMIZATION OF ALLOSTERIC HIV-1 INTEGRASE INHIBITORS DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Janet Antwi Graduate Program in Pharmaceutical Sciences The Ohio State University 2017 Dissertation Committee: Professor James R. Fuchs, Advisor Professor Werner Tjarks Professor Karl A. Werbovetz Copyright by Janet Antwi 2017 Abstract In the past quarter century, there has been tremendous progress in the discovery of antiretroviral therapy, making HIV/AIDS a manageable chronic disease. However, the HIV virus is relentless and continues to evolve under drug pressure to escape control and continue infection. The enzyme HIV integrase is responsible for the incorporation of viral double stranded DNA into a host chromosomal DNA and has recently become an attractive target in combating HIV resistance. Raltegravir (RAL), elvitegravir (EVG) and dolutegravir (DTG) are three clinically approved active-site integrase inhibitors. Unfortunately, mutations of the enzyme observed in patients have resulted in resistance to thedrug in the clinic. A new approach to targeting integrase (IN) is the development of allosteric inhibitors that specifically target the protein-protein interaction between IN and its cellular cofactor LEDGF/p75. Recently discovered quinoline-based allosteric integrase inhibitor (ALLINI) B1224436 was the first compound to advance into clinical trials but was discontinued due to poor pharmacokinetic properties including low in vivo clearance. In addition, several reports have revealed the emergence of resistance due to mutation to quinoline based ALLINIs. Applying scaffold hopping approach, several pyridine-based, thiophenes, ii pyrazoles, isoquinolines and other heteroaromatic cores have been studied as ALLINIs. -

WO 2013/164559 Al 7 November 2013 (07.11.2013) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2013/164559 Al 7 November 2013 (07.11.2013) P O P C T (51) International Patent Classification: (74) Agent: TURNER, Craig; A.A. Thornton & Co, 235 High A61K 9/16 (2006.01) A61K 31/427 (2006.01) Holborn, London WC1V 7LE (GB). A61K 9/20 (2006.01) A61K 31/513 (2006.01) (81) Designated States (unless otherwise indicated, for every (21) International Application Number: kind of national protection available): AE, AG, AL, AM, PCT/GB20 13/000 193 AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, (22) Date: International Filing DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, 3 May 20 13 (03.05.2013) HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KM, KN, KP, (25) Filing Language: English KR, KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, (26) Publication Language: English NO, NZ, OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, (30) Priority Data: RW, SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, 1380/MUM/2012 3 May 2012 (03.05.2012) IN TM, TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, 2590/MUM/2012 6 September 2012 (06.09.2012) IN ZM, ZW. -

Original Article Antiviral Activity and Tolerability of Amdoxovir with Zidovudine in a Randomized Double-Blind Placebo- Controlled Study in HIV-1-Infected Individuals
Antiviral Therapy 2010 15:185–192 (doi: 10.3851/IMP1514) Original article Antiviral activity and tolerability of amdoxovir with zidovudine in a randomized double-blind placebo- controlled study in HIV-1-infected individuals Robert L Murphy1, Nancy M Kivel2, Carlos Zala3, Claudia Ochoa3, Phillip Tharnish2, Judy Mathew 2, Maria Luz Pascual2, Raymond F Schinazi4* 1Northwestern University, Feinberg School of Medicine, Chicago, IL, USA 2RFS Pharma, LLC, Tucker, GA, USA 3Hospital Privado Modelo, Florida-Buenos Aires B1602DBG, Argentina 4Center for AIDS Research, Laboratory of Biochemical Pharmacology, Department of Pediatrics, Emory University School of Medicine and Veterans Affairs Medical Center, Atlanta, GA, USA *Corresponding author e-mail: [email protected] Background: Amdoxovir acts synergistically with zido- day 10 were determined. Laboratory and clinical safety vudine in vitro and the combination prevents or delays monitoring were performed. the selection of thymidine analogue and K65R muta- Results: Twenty-four patients were enrolled. The mean tions. In silico studies have shown that a reduced dose VL log10 change was 0.10 with placebo, -0.69 with zido- of zidovudine (200 mg) results in decreased zidovudine- vudine 200 mg, -0.55 with zidovudine 300 mg, -1.09 monophosphate levels, associated with toxicity, while with amdoxovir, -2.00 with amdoxovir plus zidovudine maintaining zidovudine-triphosphate levels, which are (200 mg) and -1.69 with amdoxovir plus zidovudine associated with antiviral effects. Here, we aimed to (300 mg). Amdoxovir plus zidovudine (200 mg) was assess the short-term tolerability and antiviral activity significantly more potent than amdoxovir monotherapy of amdoxovir in combination with reduced and standard in AUCVL and mean VL decline (P=0.019 and P=0.021, doses of zidovudine. -

Mutational Studies of Novel Screened Molecules Against Wild and Mutated HIV-1 Integrase Using Molecular Docking Studies Pawan Gupta1,2*, Prabha Garg2
Research Article Mutational studies of novel screened molecules against wild and mutated HIV-1 integrase using molecular docking studies Pawan Gupta1,2*, Prabha Garg2 ABSTRACT Background and Aim: The screened molecules which proposed novel HIV-1 integrase inhibitors were collected from the literature. Mutational studies were performed to check whether these molecules are having good binding affinity against mutated HIV-1 IN or not using molecular docking technique. Materials and Methods: First, homology models of the mutated HIV-1 IN were prepared and subsequently all the models were refined and optimized in MODELLER program. Next, molecular docking studies were performed into the active site of mutated HIV-1 IN models using the proposed inhibitors in AutoDock 4.1 program. The results of these studies were compared with the wild type docking studies. Results: The docking studies were found that some of the screened molecules (ZINC1245110, 131614, 92749, ZINC05181828, and ZINC13147504) followed the same binding patterns (in term of locations, interactions, and binding score) as found with wild type HIV-1 IN. Conclusions: Computationally, the same binding patterns were exhibited by these molecules (ZINC1245110, 131614, 92749, ZINC05181828, and ZINC13147504) against mutated models as wild type. This elucidated that these molecules having susceptibility against the drug-resistant HIV-1 IN. Hence, these molecules may be used as a starting point to design novel inhibitors against mutated HIV-1 IN, which need to be confirmed experimentally. KEY WORDS: Docking, Drug resistance, Homology modeling, HIV-1 integrase, Mutation INTRODUCTION Drug resistance is the inevitable consequence of incomplete suppression of HIV-1 replication. The Human immuno-virus (HIV) causes AIDS. -

Multi-Class Immune-Based Therap I Es Co Combination Drugs AZ T
m- IMMUNE-BASED THERAP o /R) c VIR, A LPV ) MULTI-CLASSA NFV VIR, A S T N (INDIN COMBINATION DRUGSA VIR, REZIST AZ R 754) P A ) AVX C + Inhibitors Protease RTV VIR/RITON ) CRIXIV AB A OM (KIVEXA, COMBIVIR (ZIDOVUDINE + LAMIVUDINE, AZT + 3TC) EMTRIVA Protease754, Inhibitors C VIR, (EMTRICITABINE, FTC) EPIVIR (LAMIVUDINE, 3TC) EPZICOM (KIVEXA, TPV T A PZI HE EPT (NELFIN SPD OVIR DISOPROXIL ABACAVIR + LAMIVUDINE, ABC + 3TC) RETROVIR (ZIDOVUDINE, AZT, E HIBITO F VIR, , LOPIN ZDV) TRIZIVIR (ABACAVIR + ZIDOVUDINE + LAMIVUDINE, ABC + AC A A S IR N IN TRUGGLE FOR AZT + 3TC) TRUVADA (TENOFOVIR DF + EMTRICITABINE, TDF + V ENO A ) T ABINE ( ABINE FTC) VIDEX & VIDEX EC (DIDANOSINE, DDI) VIREAD (TENOFOVIR T TV I THE (ALUVI C DISOPROXIL FUMARATE, TDF) ZERIT (STAVUDINE, D4T) ZIAGEN A (ABACAVIR, ABC) RACIVIR (RCV) AMDOXOVIR (AMDX, DAPD) ORVIR (RITON N PRI VIR, A ) IREAD ( A A V APRICITABINE (SPD754, AVX754)ELVUCITABINE (ACH- TORS ) LETR N ) A I 126,443, BETA-L-FD4C) COMBIVIR (ZIDOVUDINE + FPV A Z I LAMIVUDINE, AZT + 3TC) EMTRIVA (EMTRICITABINE, ) K A ) APTIVUS (TIPR DAPD T VIR, IPTASE IPTASE , A FTC) EPIVIR (LAMIVUDINE, 3TC) EPZICOM (KIVEXA, A PV A SQV CCESS TO ABACAVIR + LAMIVUDINE, ABC + 3TC) RETROVIR R Z ( PIVIR (LAMIVUDINE, 3TC) B A This book documents the struggle that has been faced by those E (ZIDOVUDINE, AZT, ZDV) TRIZIVIR (ABACAVIR + T AMDX VIR, VIR, A A A I STRUGGLEA requiring treatment for HIV/AIDS in India, and those affected ZIDOVUDINE + LAMIVUDINE, ABC + AZT + 3TC) MPREN AVIR + ZIDOVUDINE + LAMIVUDINE, SC EY A C TRUVADA (TENOFOVIR DF + EMTRICITABINE, by HIV/AIDS, since the first recorded incidence of HIV/AIDS in FOR R N ) TDF + FTC) VIDEX & VIDEX EC (DIDANOSINE, QUIN India in 1986. -
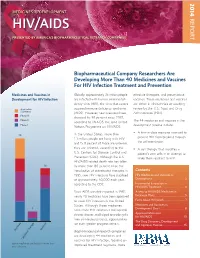
14-258 Phrma HIV/AIDS2014 0819.Indd
2014 MEDICINES IN DEVELOPMENT REPORT HIV/AIDS PRESENTED BY AMERICA’S BIOPHARMACEUTICAL RESEARCH COMPANIES Biopharmaceutical Company Researchers Are Developing More Than 40 Medicines and Vaccines For HIV Infection Treatment and Prevention Medicines and Vaccines in Globally, approximately 35 million people effective therapies, and preventative Development for HIV Infection are infected with human immunodefi - vaccines. These medicines and vaccines ciency virus (HIV), the virus that causes are either in clinical trials or awaiting Application acquired immune defi ciency syndrome review by the U.S. Food and Drug Submitted (AIDS). However, new infections have Administration (FDA). Phase III dropped by 38 percent since 2001, Phase II The 44 medicines and vaccines in the according to UNAIDS, the Joint United Phase I development pipeline include: Nations Programme on HIV/AIDS. • A fi rst-in-class medicine intended to In the United States, more than 25 prevent HIV from breaking through 1.1 million people are living with HIV the cell membrane. and 15.8 percent of those are unaware they are infected, according to the • A cell therapy that modifi es a U.S. Centers for Disease Control and patient’s own cells in an attempt to Prevention (CDC). Although the U.S. make them resistant to HIV. HIV/AIDS-related death rate has fallen 16 by more than 80 percent since the introduction of antiretroviral therapies in Contents 1995, new HIV infections have stabilized HIV Medicines and Vaccines in at approximately 50,000 each year, Development ......................................2 according to the CDC. Incremental Innovation in HIV/AIDS Treatment .......................... 4 Since AIDS was fi rst reported in 1981, Access to HIV/AIDS Medicines in nearly 40 medicines have been approved Exchange Plans ...................................5 to treat HIV infection in the United Facts About HIV/AIDS ........................7 States. -

HIV-1 Antiretroviral Drug Therapy
Downloaded from http://perspectivesinmedicine.cshlp.org/ on October 2, 2021 - Published by Cold Spring Harbor Laboratory Press HIV-1 Antiretroviral Drug Therapy Eric J. Arts1 and Daria J. Hazuda2 1Ugandan CFAR Laboratories, Division of Infectious Diseases, Department of Medicine, Case Western Reserve University, Cleveland, Ohio 44106 2Merck Research Laboratories, West Point, Pennsylvania 19486 Correspondence: [email protected]; [email protected] The most significant advance in the medical management of HIV-1 infection has been the treatment of patients with antiviral drugs, which can suppress HIV-1 replication to undetect- able levels. The discovery of HIV-1 as the causative agent of AIDS together with an ever- increasing understanding of the virus replication cycle have been instrumental in this effort by providing researchers with the knowledge and tools required to prosecute drug discovery efforts focused on targeted inhibition with specific pharmacological agents. To date, an arsenal of 24 Food and Drug Administration (FDA)-approved drugs are available for treatment of HIV-1 infections. These drugs are distributed into six distinct classes based on their molecular mechanism and resistance profiles: (1) nucleoside-analog reverse tran- scriptase inhibitors (NNRTIs), (2) non–nucleoside reverse transcriptase inhibitors (NNRTIs), (3) integrase inhibitors, (4) protease inhibitors (PIs), (5) fusion inhibitors, and (6) coreceptor antagonists. In this article, we will review the basic principles of antiretroviral drug therapy, the mode of drug action, and the factors leading to treatment failure (i.e., drug resistance). BASIC PRINCIPLES OF ANTIRETROVIRAL development and approval for human use is THERAPY described in Figure 1. Since the first HIV-1 specific antiviral drugs efore 1996, few antiretroviral treatment were given as monotherapy in the early 1990s, Boptions for HIV-1 infection existed. -

Fourth Quarter Product Sales of $2.13 Billion, up 11% Year Over Year
Gilead Sciences Announces Fourth Quarter and Full Year 2011 Financial Results February 2, 2012 4:06 PM ET - Fourth Quarter Product Sales of $2.13 Billion, up 11% Year over Year - - Full Year 2011 Product Sales of $8.10 Billion, up 10% over 2010 - - Full Year 2011 Non-GAAP EPS of $3.86, up 5% over 2010 - - Full Year 2011 Operating Cash Flows of $3.64 Billion - FOSTER CITY, Calif.--(BUSINESS WIRE)--Feb. 2, 2012-- Gilead Sciences, Inc. (Nasdaq:GILD) announced today its results of operations for the fourth quarter and full year 2011. Total revenues for the fourth quarter of 2011 increased 10 percent to $2.20 billion, from $2.00 billion for the fourth quarter of 2010. Net income for the fourth quarter of 2011 was $665.1 million, or $0.87 per diluted share, compared to $629.4 million, or $0.76 per diluted share for the fourth quarter of 2010. Non-GAAP net income for the fourth quarter of 2011, which excludes after-tax acquisition-related, restructuring and stock-based compensation expenses, was $743.1 million, or $0.97 per diluted share, compared to $779.3 million, or $0.95 per diluted share for the fourth quarter of 2010. Full year 2011 total revenues were $8.39 billion, up 5 percent compared to $7.95 billion for 2010. Net income for 2011 was $2.80 billion, or $3.55 per diluted share, compared to $2.90 billion, or $3.32 per diluted share for 2010. Non-GAAP net income for 2011, which excludes after-tax acquisition-related, restructuring and stock-based compensation expenses, was $3.04 billion, or $3.86 per diluted share, compared to $3.21 billion, or $3.69 per diluted share for 2010. -

This Project Has Been Supported with Unrestriced Grants from Abbvie Gilead Sciences HEXAL Janssen-Cilag MSD Viiv Healthcare By
This project has been supported with unrestriced grants from AbbVie Gilead Sciences HEXAL Janssen-Cilag MSD ViiV Healthcare By Marcus Altfeld, Hamburg/Boston (USA) Achim Barmeyer, Dortmund Georg Behrens, Hannover Dirk Berzow, Hamburg Christoph Boesecke, Bonn Patrick Braun, Aachen Thomas Buhk, Hamburg Rob Camp, Barcelona (Spain/USA) Rika Draenert, Munich Christian Eggers, Linz (Austria) Stefan Esser, Essen Gerd Fätkenheuer, Cologne Gunar Günther, Windhoek (Namibia) Thomas Harrer, Erlangen Christian Herzmann, Borstel Christian Hoffmann, Hamburg Heinz-August Horst, Kiel Martin Hower, Dortmund Christoph Lange, Borstel Thore Lorenzen, Hamburg Tim Niehues, Krefeld Christian Noah, Hamburg Ramona Pauli, Munich Ansgar Rieke, Koblenz Jürgen Kurt Rockstroh, Bonn Thorsten Rosenkranz, Hamburg Bernhard Schaaf, Dortmund Ulrike Sonnenberg-Schwan, Munich Christoph D. Spinner, Munich Thomas Splettstoesser (Figures), Berlin Matthias Stoll, Hannover Hendrik Streeck, Essen/Boston (USA) Jan Thoden, Freiburg Markus Unnewehr, Dortmund Mechthild Vocks-Hauck, Berlin Jan-Christian Wasmuth, Bonn Michael Weigel, Schweinfurt Thomas Weitzel, Santiago (Chile) Eva Wolf, Munich HIV 2015/16 www.hivbook.com Edited by Christian Hoffmann and Jürgen K. Rockstroh Medizin Fokus Verlag IV Christian Hoffmann, M.D., Ph.D. ICH Stadtmitte (Infektionsmedizinisches Centrum Hamburg) Glockengiesserwall 1 20095 Hamburg, Germany Phone: + 49 40 2800 4200 Fax: + 49 40 2800 42020 [email protected] Jürgen K. Rockstroh, M.D., Ph.D. Department of Medicine I University of Bonn Sigmund-Freud-Strasse 25 53105 Bonn, Germany Phone: + 49 228 287 6558 Fax: + 49 228 287 5034 [email protected] HIV Medicine is an ever-changing field. The editors and authors of HIV 2015/16 have made every effort to provide information that is accurate and complete as of the date of publication.