Safety Assessment of Miraculin Using in Silico and in Vitro Digestibility Analyses T
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Food, Food Chemistry and Gustatory Sense
Food, Food Chemistry and Gustatory Sense COOH N H María González Esguevillas MacMillan Group Meeting May 12th, 2020 Food and Food Chemistry Introduction Concepts Food any nourishing substance eaten or drunk to sustain life, provide energy and promote growth any substance containing nutrients that can be ingested by a living organism and metabolized into energy and body tissue country social act culture age pleasure election It is fundamental for our life Food and Food Chemistry Introduction Concepts Food any nourishing substance eaten or drunk to sustain life, provide energy and promote growth any substance containing nutrients that can be ingested by a living organism and metabolized into energy and body tissue OH O HO O O HO OH O O O limonin OH O O orange taste HO O O O 5-caffeoylquinic acid Coffee taste OH H iPr N O Me O O 2-decanal Capsaicinoids Coriander taste Chilli burning sensation Astray, G. EJEAFChe. 2007, 6, 1742-1763 Food and Food Chemistry Introduction Concepts Food Chemistry the study of chemical processes and interactions of all biological and non-biological components of foods biological substances areas food processing techniques de Man, J. M. Principles of Food Chemistry 1999, Springer Science Fennema, O. R. Food Chemistry. 1985, 2nd edition New York: Marcel Dekker, Inc Food and Food Chemistry Introduction Concepts Food Chemistry the study of chemical processes and interactions of all biological and non-biological components of foods biological substances areas food processing techniques carbo- hydrates water minerals lipids flavors vitamins protein food enzymes colors additive Fennema, O. R. Food Chemistry. -

Cloning of a Zinc-Binding Cysteine Proteinase Inhibitor in Citrus Vascular Tissue
J. AMER. SOC. HORT. SCI. 129(5):615–623. 2004. Cloning of a Zinc-binding Cysteine Proteinase Inhibitor in Citrus Vascular Tissue Danielle R. Ellis1 and Kathryn C. Taylor2, 3 Department of Plant Sciences, University of Arizona, 303 Forbes Building, Tucson, AZ 85721 ADDITIONAL INDEX WORDS. defense proteins, Kunitz soybean proteinase inhibitor, Citrus jambhiri ABSTRACT. A partial cDNA (cvzbp-1) was cloned based on the N-terminal sequence of a citrus (Citrus L.) vascular Zn- binding protein (CVZBP) previously isolated from vascular tissue (Taylor et al., 2002). CVZBP has homology to the Kunitz soybean proteinase inhibitor (KSPI) family. Recombinant protein produced using the cDNA clone inhibited the cysteine proteinase, papain. Metal binding capacity has not been reported for any other member of this family. CVZBP was present in leaves, stems, and roots but not seeds of all citrus species examined. However, CVZBP was present in germinating seeds after the cotyledons had turned green. Within four hrs after wounding, CVZBP was undetectable in the wounded leaf and adjacent leaves. It has been suggested that many members of the KSPI family serve a function in defense. However, the expression of the CVZBP is in direct contrast with those of KSPI members that were implicated in defense response. Though systemically regulated during wounding, we suggest that CVZBP is not a defense protein but rather may function in vascular development. Overall, the role of proteinase inhibitors (PIs) is to negatively function in the quickly expanding parenchymal tissue of this regulate proteolysis when physiologically or developmentally modifi ed stem tissue in sweet potato (Yeh et al., 1997). -

WHO Guidance on Management of Snakebites
GUIDELINES FOR THE MANAGEMENT OF SNAKEBITES 2nd Edition GUIDELINES FOR THE MANAGEMENT OF SNAKEBITES 2nd Edition 1. 2. 3. 4. ISBN 978-92-9022- © World Health Organization 2016 2nd Edition All rights reserved. Requests for publications, or for permission to reproduce or translate WHO publications, whether for sale or for noncommercial distribution, can be obtained from Publishing and Sales, World Health Organization, Regional Office for South-East Asia, Indraprastha Estate, Mahatma Gandhi Marg, New Delhi-110 002, India (fax: +91-11-23370197; e-mail: publications@ searo.who.int). The designations employed and the presentation of the material in this publication do not imply the expression of any opinion whatsoever on the part of the World Health Organization concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. Dotted lines on maps represent approximate border lines for which there may not yet be full agreement. The mention of specific companies or of certain manufacturers’ products does not imply that they are endorsed or recommended by the World Health Organization in preference to others of a similar nature that are not mentioned. Errors and omissions excepted, the names of proprietary products are distinguished by initial capital letters. All reasonable precautions have been taken by the World Health Organization to verify the information contained in this publication. However, the published material is being distributed without warranty of any kind, either expressed or implied. The responsibility for the interpretation and use of the material lies with the reader. In no event shall the World Health Organization be liable for damages arising from its use. -

Enhancing Sweetness and Stability of the Single Chain Monellin MNEI
www.nature.com/scientificreports OPEN Sweeter and stronger: enhancing sweetness and stability of the single chain monellin MNEI through Received: 08 July 2016 Accepted: 07 September 2016 molecular design Published: 23 September 2016 Serena Leone1, Andrea Pica1, Antonello Merlino1, Filomena Sannino1, Piero Andrea Temussi1,2 & Delia Picone1 Sweet proteins are a family of proteins with no structure or sequence homology, able to elicit a sweet sensation in humans through their interaction with the dimeric T1R2-T1R3 sweet receptor. In particular, monellin and its single chain derivative (MNEI) are among the sweetest proteins known to men. Starting from a careful analysis of the surface electrostatic potentials, we have designed new mutants of MNEI with enhanced sweetness. Then, we have included in the most promising variant the stabilising mutation E23Q, obtaining a construct with enhanced performances, which combines extreme sweetness to high, pH-independent, thermal stability. The resulting mutant, with a sweetness threshold of only 0.28 mg/L (25 nM) is the strongest sweetener known to date. All the new proteins have been produced and purified and the structures of the most powerful mutants have been solved by X-ray crystallography. Docking studies have then confirmed the rationale of their interaction with the human sweet receptor, hinting at a previously unpredicted role of plasticity in said interaction. Sweet proteins are a family of structurally unrelated proteins that can elicit a sweet sensation in humans. To date, eight sweet and sweet taste-modifying proteins have been identified: monellin1, thaumatin2, brazzein3, pentadin4, mabinlin5, miraculin6, neoculin7 and lysozyme8. With the sole exception of lysozyme, all sweet proteins have been purified from plants, but, besides this common feature, they share no structure or sequence homology9. -

Russell's Viper (Daboia Russelii) in Bangladesh: Its Boom and Threat To
J. Asiat. Soc. Bangladesh, Sci. 44(1): 15-22, June 2018 RUSSELL’S VIPER (DABOIA RUSSELII) IN BANGLADESH: ITS BOOM AND THREAT TO HUMAN LIFE MD. FARID AHSAN1* AND MD. ABU SAEED2 1Department of Zoology, University of Chittagong, Chittagong, Bangladesh 2 555, Kazipara, Mirpur, Dhaka-1216, Bangladesh Abstract The occurrence of Russell’s viper (Daboia russelii Shaw and Nodder 1797) in Bangladesh is century old information and its rarity was known to the wildlife biologists till 2013 but its recent booming is also causing a major threat to human life in the area. Recently it has been reported from nine districts (Dinajpur, Chapai Nawabganj, Rajshahi, Naogaon, Natore, Pabna, Rajbari, Chuadanga and Patuakhali) and old records revealed 11 districts (Nilphamari, Dinajpur, Rangpur, Chapai Nawabganj, Rajshahi, Bogra, Jessore, Satkhira, Khulna, Bagerhat and Chittagong). Thus altogether 17 out of 64 districts in Bangladesh, of which Chapai Nawabganj and Rajshahi are most affected and 20 people died due to Russell’s viper bite during 2013 to 2016. Its past and present distribution in Bangladesh and death toll of its bites have been discussed. Its booming causes have also been predicted and precautions have been recommended. Research on Russell’s viper is deemed necessary due to reemergence in deadly manner. Key words: Russell’s viper, Daboia russelii, Distribution, Boom, Panic, Death toll Introduction Two species of Russell’s viper are known to occur in this universe of which Daboia russelii (Shaw and Nodder 1797) is distributed in Pakistan, India, Nepal, Bhutan, Bangladesh and Sri Lanka (www.reptile.data-base.org); while Daboia siamensis (Smith 1917) occurs in China, Myanmar, Indonesia, Thailand, Taiwan and Cambodia (Wogan 2012). -

Venom Proteomics and Antivenom Neutralization for the Chinese
www.nature.com/scientificreports OPEN Venom proteomics and antivenom neutralization for the Chinese eastern Russell’s viper, Daboia Received: 27 September 2017 Accepted: 6 April 2018 siamensis from Guangxi and Taiwan Published: xx xx xxxx Kae Yi Tan1, Nget Hong Tan1 & Choo Hock Tan2 The eastern Russell’s viper (Daboia siamensis) causes primarily hemotoxic envenomation. Applying shotgun proteomic approach, the present study unveiled the protein complexity and geographical variation of eastern D. siamensis venoms originated from Guangxi and Taiwan. The snake venoms from the two geographical locales shared comparable expression of major proteins notwithstanding variability in their toxin proteoforms. More than 90% of total venom proteins belong to the toxin families of Kunitz-type serine protease inhibitor, phospholipase A2, C-type lectin/lectin-like protein, serine protease and metalloproteinase. Daboia siamensis Monovalent Antivenom produced in Taiwan (DsMAV-Taiwan) was immunoreactive toward the Guangxi D. siamensis venom, and efectively neutralized the venom lethality at a potency of 1.41 mg venom per ml antivenom. This was corroborated by the antivenom efective neutralization against the venom procoagulant (ED = 0.044 ± 0.002 µl, 2.03 ± 0.12 mg/ml) and hemorrhagic (ED50 = 0.871 ± 0.159 µl, 7.85 ± 3.70 mg/ ml) efects. The hetero-specifc Chinese pit viper antivenoms i.e. Deinagkistrodon acutus Monovalent Antivenom and Gloydius brevicaudus Monovalent Antivenom showed negligible immunoreactivity and poor neutralization against the Guangxi D. siamensis venom. The fndings suggest the need for improving treatment of D. siamensis envenomation in the region through the production and the use of appropriate antivenom. Daboia is a genus of the Viperinae subfamily (family: Viperidae), comprising a group of vipers commonly known as Russell’s viper native to the Old World1. -
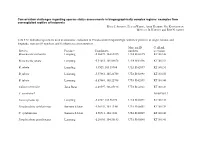
Conservation Challenges Regarding Species Status Assessments in Biogeographically Complex Regions: Examples from Overexploited Reptiles of Indonesia KYLE J
Conservation challenges regarding species status assessments in biogeographically complex regions: examples from overexploited reptiles of Indonesia KYLE J. SHANEY, ELIJAH WOSTL, AMIR HAMIDY, NIA KURNIAWAN MICHAEL B. HARVEY and ERIC N. SMITH TABLE S1 Individual specimens used in taxonomic evaluation of Pseudocalotes tympanistriga, with their province of origin, latitude and longitude, museum ID numbers, and GenBank accession numbers. Museum ID GenBank Species Province Coordinates numbers accession Bronchocela cristatella Lampung -5.36079, 104.63215 UTA R 62895 KT180148 Bronchocela jubata Lampung -5.54653, 105.04678 UTA R 62896 KT180152 B. jubata Lampung -5.5525, 105.18384 UTA R 62897 KT180151 B. jubata Lampung -5.57861, 105.22708 UTA R 62898 KT180150 B. jubata Lampung -5.57861, 105.22708 UTA R 62899 KT180146 Calotes versicolor Jawa Barat -6.49597, 106.85198 UTA R 62861 KT180149 C. versicolor* NC009683.1 Gonocephalus sp. Lampung -5.2787, 104.56198 UTA R 60571 KT180144 Pseudocalotes cybelidermus Sumatra Selatan -4.90149, 104.13401 UTA R 60551 KT180139 P. cybelidermus Sumatra Selatan -4.90711, 104.1348 UTA R 60549 KT180140 Pseudocalotes guttalineatus Lampung -5.28105, 104.56183 UTA R 60540 KT180141 P. guttalineatus Sumatra Selatan -4.90681, 104.13457 UTA R 60501 KT180142 Pseudocalotes rhammanotus Lampung -4.9394, 103.85292 MZB 10804 KT180147 Pseudocalotes species 4 Sumatra Barat -2.04294, 101.31129 MZB 13295 KT211019 Pseudocalotes tympanistriga Jawa Barat -6.74181, 107.0061 UTA R 60544 KT180143 P. tympanistriga Jawa Barat -6.74181, 107.0061 UTA R 60547 KT180145 Pogona vitticeps* AB166795.1 *Entry to GenBank by previous authors TABLE S2 Reptile species currently believed to occur Java and Sumatra, Indonesia, with IUCN Red List status, and certainty of occurrence. -

Crystal Structure of Crataeva Tapia Bark Protein (Cratabl) and Its Effect in Human Prostate Cancer Cell Lines
Crystal Structure of Crataeva tapia Bark Protein (CrataBL) and Its Effect in Human Prostate Cancer Cell Lines Rodrigo da Silva Ferreira1., Dongwen Zhou2., Joana Gasperazzo Ferreira1, Mariana Cristina Cabral Silva1, Rosemeire Aparecida Silva-Lucca3, Reinhard Mentele4,5, Edgar Julian Paredes-Gamero1, Thiago Carlos Bertolin6, Maria Tereza dos Santos Correia7, Patrı´cia Maria Guedes Paiva7, Alla Gustchina2, Alexander Wlodawer2*, Maria Luiza Vilela Oliva1* 1 Departamento de Bioquı´mica, Universidade Federal de Sa˜o Paulo, Sa˜o Paulo, Sa˜o Paulo, Brazil, 2 Macromolecular Crystallography Laboratory, Center for Cancer Research, National Cancer Institute, Frederick, Maryland, United States of America, 3 Centro de Engenharias e Cieˆncias Exatas, Universidade Estadual do Oeste do Parana´, Toledo, Parana´, Brazil, 4 Institute of Clinical Neuroimmunology LMU, Max-Planck-Institute for Biochemistry, Martinsried, Munich, Germany, 5 Department for Protein Analytics, Max-Planck-Institute for Biochemistry, Martinsried, Munich, Germany, 6 Departamento de Biofı´sica, Universidade Federal de Sa˜o Paulo, Sa˜o Paulo, Sa˜o Paulo, Brazil, 7 Departamento de Bioquı´mica, Universidade Federal de Pernambuco, Recife, Pernambuco, Brazil Abstract A protein isolated from the bark of Crataeva tapia (CrataBL) is both a Kunitz-type plant protease inhibitor and a lectin. We have determined the amino acid sequence and three-dimensional structure of CrataBL, as well as characterized its selected biochemical and biological properties. We found two different isoforms of CrataBL isolated from the original source, differing in positions 31 (Pro/Leu); 92 (Ser/Leu); 93 (Ile/Thr); 95 (Arg/Gly) and 97 (Leu/Ser). CrataBL showed relatively weak inhibitory activity against trypsin (Kiapp =43 mM) and was more potent against Factor Xa (Kiapp = 8.6 mM), but was not active against a number of other proteases. -

P. 1 AC27 Inf. 7 (English Only / Únicamente En Inglés / Seulement
AC27 Inf. 7 (English only / únicamente en inglés / seulement en anglais) CONVENTION ON INTERNATIONAL TRADE IN ENDANGERED SPECIES OF WILD FAUNA AND FLORA ____________ Twenty-seventh meeting of the Animals Committee Veracruz (Mexico), 28 April – 3 May 2014 Species trade and conservation IUCN RED LIST ASSESSMENTS OF ASIAN SNAKE SPECIES [DECISION 16.104] 1. The attached information document has been submitted by IUCN (International Union for Conservation of * Nature) . It related to agenda item 19. * The geographical designations employed in this document do not imply the expression of any opinion whatsoever on the part of the CITES Secretariat or the United Nations Environment Programme concerning the legal status of any country, territory, or area, or concerning the delimitation of its frontiers or boundaries. The responsibility for the contents of the document rests exclusively with its author. AC27 Inf. 7 – p. 1 Global Species Programme Tel. +44 (0) 1223 277 966 219c Huntingdon Road Fax +44 (0) 1223 277 845 Cambridge CB3 ODL www.iucn.org United Kingdom IUCN Red List assessments of Asian snake species [Decision 16.104] 1. Introduction 2 2. Summary of published IUCN Red List assessments 3 a. Threats 3 b. Use and Trade 5 c. Overlap between international trade and intentional use being a threat 7 3. Further details on species for which international trade is a potential concern 8 a. Species accounts of threatened and Near Threatened species 8 i. Euprepiophis perlacea – Sichuan Rat Snake 9 ii. Orthriophis moellendorfi – Moellendorff's Trinket Snake 9 iii. Bungarus slowinskii – Red River Krait 10 iv. Laticauda semifasciata – Chinese Sea Snake 10 v. -

Aminobutyric Acid Priming Acquisition and Defense Response of Mango Fruit to Colletotrichum Gloeosporioides Infection Based on Quantitative Proteomics
Article β-Aminobutyric Acid Priming Acquisition and Defense Response of Mango Fruit to Colletotrichum gloeosporioides Infection Based on Quantitative Proteomics Taotao Li 1, Panhui Fan 2, Ze Yun 1 , Guoxiang Jiang 1, Zhengke Zhang 1,2,* and Yueming Jiang 1 1 Key Laboratory of Plant Resources Conservation and Sustainable Utilization/Guangdong Provincial Key Laboratory of Applied Botany, South China Botanical Garden, Chinese Academy of Sciences, Guangzhou 510650, China 2 College of Food Science and Engineering, Hainan University, Haikou 570228, China * Correspondence: [email protected] Received: 2 August 2019; Accepted: 2 September 2019; Published: 4 September 2019 Abstract: β-aminobutyric acid (BABA) is a new environmentally friendly agent to induce disease resistance by priming of defense in plants. However, molecular mechanisms underlying BABA-induced priming defense are not fully understood. Here, comprehensive analysis of priming mechanism of BABA-induced resistance was investigated based on mango-Colletotrichum gloeosporioides interaction system using iTRAQ-based proteome approach. Results showed that BABA treatments effectively inhibited the expansion of anthracnose caused by C. gleosporioides in mango fruit. Proteomic results revealed that stronger response to pathogen in BABA-primed mango fruit after C. gleosporioides inoculation might be attributed to differentially accumulated proteins involved in secondary metabolism, defense signaling and response, transcriptional regulation, protein post-translational modification, etc. Additionally, we testified the involvement of non-specific lipid-transfer protein (nsLTP) in the priming acquisition at early priming stage and memory in BABA-primed mango fruit. Meanwhile, spring effect was found in the primed mango fruit, indicated by inhibition of defense-related proteins at priming phase but stronger activation of defense response when exposure to pathogen compared with non-primed fruit. -

The Primary Structure of Inhibitor of Cysteine Proteinases from Potato
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Volume 333, number 1,2, 15-20 FEBS 13125 October 1993 0 1993 Federation of European Biochemical Societies 00145793/93/%6.00 The primary structure of inhibitor of cysteine proteinases from potato I. Kriiaj*, M. DrobniE-KoSorok, J. Brzin, R. Jerala, V. Turk Department of Biochemistry and Molecular Biology, Joief Stefan Institute, Jamova 39, 61111 Ljubljana, Slovenia Received 26 July 1993; revised version received 3 September 1993 The complete amino acid sequence of the cysteine proteinase inhibitor from potato tubers was determined. The inhibitor is a single-chain protein having 180 amino acid residues. Its primary structure was elucidated by automatic degradation of the intact protein and sequence analysis of peptides generated by CNBr, trypsin and glycyl endopeptidase. A search through the protein sequence database showed homology to other plant proteinase inhibitors of different specificities and non-inhibitory proteins of M, around 20,000. On the basis of sequence homology, prediction of secondary structure and fold compatibility, based on a 3DlD score to the threedimensional profile of Erythrina caffra trypsin inhibitor, we suggest that the potato cysteine proteinase inhibitor belongs to the superfamily of proteins that have the same pattern of three-dimensional structure as soybean trypsin inhibitor. This superfamily would therefore include proteins that inhibit three different classes of proteinases - serine, cysteine and aspartic proteinases. Cysteine proteinase inhibitor; Amino acid sequence; Solunum tuberown; Soybean trypsin inhibitor superfamily 1. INTRODUCTION ized as a potent inhibitor of lysosomal proteinase cathepsin L with a Ki 0.07 nM [9]. -

Modeling to Understand Plant Protein Structure-Function Relationships—Implications for Seed Storage Proteins
molecules Review Modeling to Understand Plant Protein Structure-Function Relationships—Implications for Seed Storage Proteins 1,2, 1, 2 1, Faiza Rasheed y, Joel Markgren y , Mikael Hedenqvist and Eva Johansson * 1 Department of Plant Breeding, The Swedish University of Agricultural Sciences, Box 101, SE-230 53 Alnarp, Sweden; [email protected] (F.R.); [email protected] (J.M.) 2 School of Chemical Science and Engineering, Fibre and Polymer Technology, KTH Royal Institute of Technology, SE–100 44 Stockholm, Sweden; [email protected] * Correspondence: [email protected] These authors contributed equally to this work. y Received: 2 January 2020; Accepted: 14 February 2020; Published: 17 February 2020 Abstract: Proteins are among the most important molecules on Earth. Their structure and aggregation behavior are key to their functionality in living organisms and in protein-rich products. Innovations, such as increased computer size and power, together with novel simulation tools have improved our understanding of protein structure-function relationships. This review focuses on various proteins present in plants and modeling tools that can be applied to better understand protein structures and their relationship to functionality, with particular emphasis on plant storage proteins. Modeling of plant proteins is increasing, but less than 9% of deposits in the Research Collaboratory for Structural Bioinformatics Protein Data Bank come from plant proteins. Although, similar tools are applied as in other proteins, modeling of plant proteins is lagging behind and innovative methods are rarely used. Molecular dynamics and molecular docking are commonly used to evaluate differences in forms or mutants, and the impact on functionality.