In Situ Identification of Keratinhydrolyzing Organisms In
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Taxonomy of the Azotobacteraceae Determined by Using Immunoelectrophoresis Y
INTERNATIONALJOURNAL OF SYSTEMATICBACTERIOLOGY, Apr. 1983, p. 147-156 Vol. 33, No. 2 0020-7713/83/020147-10$02. WO Copyright 0 1983, International Union of Microbiological Societies a Taxonomy of the Azotobacteraceae Determined by Using Immunoelectrophoresis Y. T. TCHAN,'* Z. WYSZOMIRSKA-DREHER,' P. B. NEW,' AND J.-C. ZHOU' Department of Microbiology, University of Sydney, New South Wales, 2006, Australia, and Hua-ckung Agricultural College, Wuhan, People's Republic of China' The similarities of various strains of Azotobacter spp. and Azomonas spp. to reference strains of Azotobacter paspali, Azotobacter vinelandii, Azotobacter chroococcum, Azomonas agilis, Azomonas insignis, and Azomonas macrocyto- genes were determined by rocket line immunoelectrophoresis. The strains of Azotobacter paspali and Azotobacter vinelandii used were immunologically more homogeneous than the strains of Azotobacter chroococcum studied, possibly due to the more diverse geographical origins of the Azotobacter chroococcum strains. Low values were obtained for the mean immunological distances (1 - proportion of immunoprecipitation bands shared between strains) between Azotobacter paspali and Azotobacter vinelandii strains, suggesting that these two species are immunologically closely related. Immunological distances from the Azotobacter chroococcum reference strain were similar for Azotobacter paspali and for other undisputed members of the genus Azotobacter, which makes it reasonable to retain Azotobacter paspali in this genus. When the three Azotobacter antisera were used, all Azotobacter species had mean immunological distances of less than 0.5, whereas the Azomonas species were immunologically more distant , showing that the six species of Azotobacter form an immunologically related group which is distinct from the Azomonas species. Our results with the three Azomonas antisera show that each species of Azoinonas is immunologically distant from the other species, as well as from the Azotobacter species. -

Bourbon Gumbo” 10/13/2016
“Bourbon Gumbo” 10/13/2016 Microbial Analysis Report Table of Contents Executive Summary ----------------------------------------------------------------------------------------------------------------2 Background ---------------------------------------------------------------------------------------------------------------------2 Results ---------------------------------------------------------------------------------------------------------------------------2 Coliforms ------------------------------------------------------------------------------------------------------------------------4 Non-Coliforms that can trigger Coliform test ----------------------------------------------------------------------------4 Fecal Indicator Bacteria -------------------------------------------------------------------------------------------------------4 Potential Pathogens ------------------------------------------------------------------------------------------------------------4 Freshwater or Marine Bacteria (potential sign of surface water intrusion) -------------------------------------------4 Nitrogen Fixing Bacteria ------------------------------------------------------------------------------------------------------5 Carbon Fixing Bacteria --------------------------------------------------------------------------------------------------------5 Ammonia Oxidizing Bacteria ------------------------------------------------------------------------------------------------5 Nitrite Oxidizing Bacteria ----------------------------------------------------------------------------------------------------5 -

Core Sulphate-Reducing Microorganisms in Metal-Removing Semi-Passive Biochemical Reactors and the Co-Occurrence of Methanogens
microorganisms Article Core Sulphate-Reducing Microorganisms in Metal-Removing Semi-Passive Biochemical Reactors and the Co-Occurrence of Methanogens Maryam Rezadehbashi and Susan A. Baldwin * Chemical and Biological Engineering, University of British Columbia, 2360 East Mall, Vancouver, BC V6T 1Z3, Canada; [email protected] * Correspondence: [email protected]; Tel.: +1-604-822-1973 Received: 2 January 2018; Accepted: 17 February 2018; Published: 23 February 2018 Abstract: Biochemical reactors (BCRs) based on the stimulation of sulphate-reducing microorganisms (SRM) are emerging semi-passive remediation technologies for treatment of mine-influenced water. Their successful removal of metals and sulphate has been proven at the pilot-scale, but little is known about the types of SRM that grow in these systems and whether they are diverse or restricted to particular phylogenetic or taxonomic groups. A phylogenetic study of four established pilot-scale BCRs on three different mine sites compared the diversity of SRM growing in them. The mine sites were geographically distant from each other, nevertheless the BCRs selected for similar SRM types. Clostridia SRM related to Desulfosporosinus spp. known to be tolerant to high concentrations of copper were members of the core microbial community. Members of the SRM family Desulfobacteraceae were dominant, particularly those related to Desulfatirhabdium butyrativorans. Methanogens were dominant archaea and possibly were present at higher relative abundances than SRM in some BCRs. Both hydrogenotrophic and acetoclastic types were present. There were no strong negative or positive co-occurrence correlations of methanogen and SRM taxa. Knowing which SRM inhabit successfully operating BCRs allows practitioners to target these phylogenetic groups when selecting inoculum for future operations. -

Macellibacteroides Fermentans Gen. Nov., Sp. Nov., a Member of the Family Porphyromonadaceae Isolated from an Upflow Anaerobic Filter Treating Abattoir Wastewaters
International Journal of Systematic and Evolutionary Microbiology (2012), 62, 2522–2527 DOI 10.1099/ijs.0.032508-0 Macellibacteroides fermentans gen. nov., sp. nov., a member of the family Porphyromonadaceae isolated from an upflow anaerobic filter treating abattoir wastewaters Linda Jabari,1,2 Hana Gannoun,2 Jean-Luc Cayol,1 Abdeljabbar Hedi,1 Mitsuo Sakamoto,3 Enevold Falsen,4 Moriya Ohkuma,3 Moktar Hamdi,2 Guy Fauque,1 Bernard Ollivier1 and Marie-Laure Fardeau1 Correspondence 1Aix-Marseille Universite´ du Sud Toulon-Var, CNRS/INSU, IRD, MIO, UM 110, Case 925, Marie-Laure Fardeau 163 Avenue de Luminy, 13288 Marseille Cedex 9, France [email protected] 2Laboratoire d’Ecologie et de Technologie Microbienne, Institut National des Sciences Applique´es et de Technologie, Centre Urbain Nord, BP 676, 1080 Tunis Cedex, Tunisia 3Microbe Division/Japan Collection of Microorganisms, RIKEN BioResource Center 2-1 Hirosawa, Wako, Saitama 351-0198, Japan 4CCUG, Culture Collection, Department of Clinical Bacteriology, University of Go¨teborg, 41346 Go¨teborg, Sweden A novel obligately anaerobic, non-spore-forming, rod-shaped mesophilic bacterium, which stained Gram-positive but showed the typical cell wall structure of Gram-negative bacteria, was isolated from an upflow anaerobic filter treating abattoir wastewaters in Tunisia. The strain, designated LIND7HT, grew at 20–45 6C (optimum 35–40 6C) and at pH 5.0–8.5 (optimum pH 6.5–7.5). It did not require NaCl for growth, but was able to grow in the presence of up to 2 % NaCl. Sulfate, thiosulfate, elemental sulfur, sulfite, nitrate and nitrite were not used as terminal electron acceptors. -

Fatty Acid Diets: Regulation of Gut Microbiota Composition and Obesity and Its Related Metabolic Dysbiosis
International Journal of Molecular Sciences Review Fatty Acid Diets: Regulation of Gut Microbiota Composition and Obesity and Its Related Metabolic Dysbiosis David Johane Machate 1, Priscila Silva Figueiredo 2 , Gabriela Marcelino 2 , Rita de Cássia Avellaneda Guimarães 2,*, Priscila Aiko Hiane 2 , Danielle Bogo 2, Verônica Assalin Zorgetto Pinheiro 2, Lincoln Carlos Silva de Oliveira 3 and Arnildo Pott 1 1 Graduate Program in Biotechnology and Biodiversity in the Central-West Region of Brazil, Federal University of Mato Grosso do Sul, Campo Grande 79079-900, Brazil; [email protected] (D.J.M.); [email protected] (A.P.) 2 Graduate Program in Health and Development in the Central-West Region of Brazil, Federal University of Mato Grosso do Sul, Campo Grande 79079-900, Brazil; pri.fi[email protected] (P.S.F.); [email protected] (G.M.); [email protected] (P.A.H.); [email protected] (D.B.); [email protected] (V.A.Z.P.) 3 Chemistry Institute, Federal University of Mato Grosso do Sul, Campo Grande 79079-900, Brazil; [email protected] * Correspondence: [email protected]; Tel.: +55-67-3345-7416 Received: 9 March 2020; Accepted: 27 March 2020; Published: 8 June 2020 Abstract: Long-term high-fat dietary intake plays a crucial role in the composition of gut microbiota in animal models and human subjects, which affect directly short-chain fatty acid (SCFA) production and host health. This review aims to highlight the interplay of fatty acid (FA) intake and gut microbiota composition and its interaction with hosts in health promotion and obesity prevention and its related metabolic dysbiosis. -

Effect of Ph and Temperature on Microbial Community Structure And
Calicioglu et al. Biotechnol Biofuels (2018) 11:275 https://doi.org/10.1186/s13068-018-1278-6 Biotechnology for Biofuels RESEARCH Open Access Efect of pH and temperature on microbial community structure and carboxylic acid yield during the acidogenic digestion of duckweed Ozgul Calicioglu1, Michael J. Shreve1, Tom L. Richard2 and Rachel A. Brennan1* Abstract Background: Duckweeds (Lemnaceae) are efcient aquatic plants for wastewater treatment due to their high nutri- ent-uptake capabilities and resilience to severe environmental conditions. Combined with their rapid growth rates, high starch, and low lignin contents, duckweeds have also gained popularity as a biofuel feedstock for thermochemi- cal conversion and alcohol fermentation. However, studies on the acidogenic anaerobic digestion of duckweed into carboxylic acids, another group of chemicals which are precursors of higher-value chemicals and biofuels, are lacking. In this study, a series of laboratory batch experiments were performed to determine the favorable operating condi- tions (i.e., temperature and pH) to maximize carboxylic acid production from wastewater-derived duckweed during acidogenic digestion. Batch reactors with 25 g/l solid loading were operated anaerobically for 21 days under meso- philic (35 °C) or thermophilic (55 °C) conditions at an acidic (5.3) or basic (9.2) pH. At the conclusion of the experiment, the dominant microbial communities under various operating conditions were assessed using high-throughput sequencing. Results: The highest duckweed–carboxylic acid conversion of 388 28 mg acetic acid equivalent per gram volatile solids was observed under mesophilic and basic conditions, with an± average production rate of 0.59 g/l/day. This result is comparable to those reported for acidogenic digestion of other organics such as food waste. -

Comparative Genomics of the Genus Porphyromonas Identifies Adaptations for Heme Synthesis Within the Prevalent Canine Oral Species Porphyromonas Cangingivalis
GBE Comparative Genomics of the Genus Porphyromonas Identifies Adaptations for Heme Synthesis within the Prevalent Canine Oral Species Porphyromonas cangingivalis Ciaran O’Flynn1,*, Oliver Deusch1, Aaron E. Darling2, Jonathan A. Eisen3,4,5, Corrin Wallis1,IanJ.Davis1,and Stephen J. Harris1 1 The WALTHAM Centre for Pet Nutrition, Waltham-on-the-Wolds, United Kingdom Downloaded from 2The ithree Institute, University of Technology Sydney, Ultimo, New South Wales, Australia 3Department of Evolution and Ecology, University of California, Davis 4Department of Medical Microbiology and Immunology, University of California, Davis 5UC Davis Genome Center, University of California, Davis http://gbe.oxfordjournals.org/ *Corresponding author: E-mail: ciaran.ofl[email protected]. Accepted: November 6, 2015 Abstract Porphyromonads play an important role in human periodontal disease and recently have been shown to be highly prevalent in canine mouths. Porphyromonas cangingivalis is the most prevalent canine oral bacterial species in both plaque from healthy gingiva and at University of Technology, Sydney on January 17, 2016 plaque from dogs with early periodontitis. The ability of P. cangingivalis to flourish in the different environmental conditions char- acterized by these two states suggests a degree of metabolic flexibility. To characterize the genes responsible for this, the genomes of 32 isolates (including 18 newly sequenced and assembled) from 18 Porphyromonad species from dogs, humans, and other mammals were compared. Phylogenetic trees inferred using core genes largely matched previous findings; however, comparative genomic analysis identified several genes and pathways relating to heme synthesis that were present in P. cangingivalis but not in other Porphyromonads. Porphyromonas cangingivalis has a complete protoporphyrin IX synthesis pathway potentially allowing it to syn- thesize its own heme unlike pathogenic Porphyromonads such as Porphyromonas gingivalis that acquire heme predominantly from blood. -

Supporting Information
Supporting Information Lozupone et al. 10.1073/pnas.0807339105 SI Methods nococcus, and Eubacterium grouped with members of other Determining the Environmental Distribution of Sequenced Genomes. named genera with high bootstrap support (Fig. 1A). One To obtain information on the lifestyle of the isolate and its reported member of the Bacteroidetes (Bacteroides capillosus) source, we looked at descriptive information from NCBI grouped firmly within the Firmicutes. This taxonomic error was (www.ncbi.nlm.nih.gov/genomes/lproks.cgi) and other related not surprising because gut isolates have often been classified as publications. We also determined which 16S rRNA-based envi- Bacteroides based on an obligate anaerobe, Gram-negative, ronmental surveys of microbial assemblages deposited near- nonsporulating phenotype alone (6, 7). A more recent 16S identical sequences in GenBank. We first downloaded the gbenv rRNA-based analysis of the genus Clostridium defined phylo- files from the NCBI ftp site on December 31, 2007, and used genetically related clusters (4, 5), and these designations were them to create a BLAST database. These files contain GenBank supported in our phylogenetic analysis of the Clostridium species in the HGMI pipeline. We thus designated these Clostridium records for the ENV database, a component of the nonredun- species, along with the species from other named genera that dant nucleotide database (nt) where 16S rRNA environmental cluster with them in bootstrap supported nodes, as being within survey data are deposited. GenBank records for hits with Ͼ98% these clusters. sequence identity over 400 bp to the 16S rRNA sequence of each of the 67 genomes were parsed to get a list of study titles Annotation of GTs and GHs. -

Crystalline Iron Oxides Stimulate Methanogenic Benzoate Degradation in Marine Sediment-Derived Enrichment Cultures
The ISME Journal https://doi.org/10.1038/s41396-020-00824-7 ARTICLE Crystalline iron oxides stimulate methanogenic benzoate degradation in marine sediment-derived enrichment cultures 1,2 1 3 1,2 1 David A. Aromokeye ● Oluwatobi E. Oni ● Jan Tebben ● Xiuran Yin ● Tim Richter-Heitmann ● 2,4 1 5 5 1 2,3 Jenny Wendt ● Rolf Nimzyk ● Sten Littmann ● Daniela Tienken ● Ajinkya C. Kulkarni ● Susann Henkel ● 2,4 2,4 1,3 2,3,4 1,2 Kai-Uwe Hinrichs ● Marcus Elvert ● Tilmann Harder ● Sabine Kasten ● Michael W. Friedrich Received: 19 May 2020 / Revised: 9 October 2020 / Accepted: 22 October 2020 © The Author(s) 2020. This article is published with open access Abstract Elevated dissolved iron concentrations in the methanic zone are typical geochemical signatures of rapidly accumulating marine sediments. These sediments are often characterized by co-burial of iron oxides with recalcitrant aromatic organic matter of terrigenous origin. Thus far, iron oxides are predicted to either impede organic matter degradation, aiding its preservation, or identified to enhance organic carbon oxidation via direct electron transfer. Here, we investigated the effect of various iron oxide phases with differing crystallinity (magnetite, hematite, and lepidocrocite) during microbial 1234567890();,: 1234567890();,: degradation of the aromatic model compound benzoate in methanic sediments. In slurry incubations with magnetite or hematite, concurrent iron reduction, and methanogenesis were stimulated during accelerated benzoate degradation with methanogenesis as the dominant electron sink. In contrast, with lepidocrocite, benzoate degradation, and methanogenesis were inhibited. These observations were reproducible in sediment-free enrichments, even after five successive transfers. Genes involved in the complete degradation of benzoate were identified in multiple metagenome assembled genomes. -

Significance of Donor Human Milk
fmicb-09-01376 June 26, 2018 Time: 17:31 # 1 ORIGINAL RESEARCH published: 27 June 2018 doi: 10.3389/fmicb.2018.01376 Preterm Gut Microbiome Depending on Feeding Type: Significance of Donor Human Milk Anna Parra-Llorca1, María Gormaz1,2, Cristina Alcántara3, María Cernada1,2, Antonio Nuñez-Ramiro1,2, Máximo Vento1,2*† and Maria C. Collado3*† 1 Neonatal Research Group, Health Research Institute La Fe, University and Polytechnic Hospital La Fe, Valencia, Spain, 2 Division of Neonatology, University and Polytechnic Hospital La Fe, Valencia, Spain, 3 Department of Biotechnology, Institute of Agrochemistry and Food Technology, Spanish National Research Council, Valencia, Spain Preterm microbial colonization is affected by gestational age, antibiotic treatment, type of birth, but also by type of feeding. Breast milk has been acknowledged as the gold standard for human nutrition. In preterm infants breast milk has been associated with improved growth and cognitive development and a reduced risk of necrotizing enterocolitis and late onset sepsis. In the absence of their mother’s own milk (MOM), pasteurized donor human milk (DHM) could be the best available alternative due to its similarity to the former. However, little is known about the effect of DHM upon preterm Edited by: Sandra Torriani, microbiota and potential biological implications. Our objective was to determine the University of Verona, Italy impact of DHM upon preterm gut microbiota admitted in a referral neonatal intensive Reviewed by: care unit (NICU). A prospective observational cohort study in NICU of 69 neonates Carlotta De Filippo, <32 weeks of gestation and with a birth weight ≤1,500 g was conducted. -
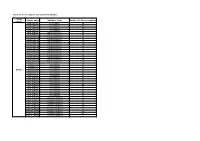
Table S1: List of Samples Included in the Analysis
Table S1: list of samples included in the analysis Study Sample name Inhibitory status Number of days at sampling number DNA.0P2T4 No inhibition 29 DNA.0P2T6 No inhibition 57 DNA.10P2T4 No inhibition 29 DNA.10P2T6 No inhibition 57 DNA.75P2T4 Phenol inhibition 29 DNA.75P2T6 Phenol inhibition 57 DNA.100P2T4 Phenol inhibition 29 DNA.100P2T6 Phenol inhibition 57 DNA.125P1T4 Phenol inhibition 29 DNA.125P1T6 Phenol inhibition 57 DNA.125P2T4 Phenol inhibition 29 DNA.125P2T6 Phenol inhibition 57 DNA.125P3T4 Phenol inhibition 29 DNA.125P3T6 Phenol inhibition 57 DNA.150P2T4 Phenol inhibition 29 DNA.150P2T6 Phenol inhibition 57 DNA.200P2T4 Phenol inhibition 29 DNA.200P2T6 Phenol inhibition 57 DNA.0N2T4 No inhibition 29 DNA.0N2T5 No inhibition 42 DNA.0N2T6 No inhibition 57 Study 1 DNA.5N2T4 No inhibition 29 DNA.5N2T5 No inhibition 42 DNA.5N2T6 No inhibition 57 DNA.10N2T4 No inhibition 29 DNA.10N2T5 No inhibition 42 DNA.10N2T6 No inhibition 57 DNA.15N2T4 No inhibition 29 DNA.15N2T5 No inhibition 42 DNA.15N2T6 No inhibition 57 DNA.25N2T4 No inhibition 29 DNA.25N2T5 No inhibition 42 DNA.25N2T6 No inhibition 57 DNA.75N2T4 Ammonia inhibition 29 DNA.75N2T5 Ammonia inhibition 42 DNA.75N2T6 Ammonia inhibition 57 DNA.100N2T4 Ammonia inhibition 29 DNA.100N2T5 Ammonia inhibition 42 DNA.100N2T6 Ammonia inhibition 57 DNA.250N2T4 Ammonia inhibition 29 DNA.250N2T5 Ammonia inhibition 42 DNA.250N2T6 Ammonia inhibition 57 nono2T3 No inhibition 16 noN2T4 Ammonia inhibition 23 noN2T8 Ammonia inhibition 60 noN2T9 Ammonia inhibition 85 noPhi2T4 Phenol inhibition 23 noPhi2T5 -

Dehalobacter Restrictus PER-K23T
Standards in Genomic Sciences (2013) 8:375-388 DOI:10.4056/sigs.3787426 Complete genome sequence of Dehalobacter restrictus T PER-K23 Thomas Kruse1*, Julien Maillard2, Lynne Goodwin3,4, Tanja Woyke3, Hazuki Teshima3,4, David Bruce3,4, Chris Detter3,4, Roxanne Tapia3,4, Cliff Han3,4, Marcel Huntemann3, Chia-Lin Wei3, James Han3, Amy Chen3, Nikos Kyrpides3, Ernest Szeto3, Victor Markowitz3, Natalia Ivanova3, Ioanna Pagani3, Amrita Pati3, Sam Pitluck3, Matt Nolan3, Christof Holliger2, and Hauke Smidt1 1 Wageningen University, Agrotechnology and Food Sciences, Laboratory of Microbiology, Dreijenplein 10, NL-6703 HB Wageningen, The Netherlands. 2 Ecole Polytechnique Fédérale de Lausanne (EPFL), School of Architecture, Civil and Environmental Engineering, Laboratory for Environmental Biotechnology, Station 6, CH- 1015 Lausanne, Switzerland. 3 DOE Joint Genome Institute, Walnut Creek, California, USA 4 Los Alamos National Laboratory, Bioscience Division, Los Alamos, New Mexico, USA *Corresponding author: Thomas Kruse ([email protected]) Keywords: Dehalobacter restrictus type strain, anaerobe, organohalide respiration, PCE, TCE, reductive dehalogenases Dehalobacter restrictus strain PER-K23 (DSM 9455) is the type strain of the species Dehalobacter restrictus. D. restrictus strain PER-K23 grows by organohalide respiration, cou- pling the oxidation of H2 to the reductive dechlorination of tetra- or trichloroethene. Growth has not been observed with any other electron donor or acceptor, nor has fermentative growth been shown. Here we introduce the first full genome of a pure culture within the ge- nus Dehalobacter. The 2,943,336 bp long genome contains 2,826 protein coding and 82 RNA genes, including 5 16S rRNA genes. Interestingly, the genome contains 25 predicted re- ductive dehalogenase genes, the majority of which appear to be full length.