ENZYMOLOGY of DNA REPLICATION - I
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

(NAP) and Illustration of DNA Flexure Angles at Single Molecule Resolution Debayan Purkait†A, Debolina Bandyopadhyay†A, and Padmaja P
bioRxiv preprint doi: https://doi.org/10.1101/2020.09.11.293639; this version posted September 11, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Vital Insights into Prokaryotic Genome Compaction by Nucleoid-Associated Protein (NAP) and Illustration of DNA Flexure Angles at Single Molecule Resolution Debayan Purkait†a, Debolina Bandyopadhyay†a, and Padmaja P. Mishra*a aChemical Sciences Division, Saha Institute of Nuclear Physics, India aHomi Bhaba National Institute (HBNI), India †Both the authors have contributed equally. * Corresponding author Abstract Integration Host Factor (IHF) is a heterodimeric site-specific nucleoid-associated protein (NAP) well known for its DNA bending ability. The binding is mediated through the narrow minor grooves of the consensus sequence, involving van der-Waals interaction and hydrogen bonding. Although the DNA bend state of IHF has been captured by both X-ray Crystallography and Atomic Force Microscopy (AFM), the range of flexibility and degree of heterogeneity in terms of quantitative analysis of the nucleoprotein complex has largely remained unexplored. Here we have monitored and compared the trajectories of the conformational dynamics of a dsDNA upon binding of wild-type (wt) and single-chain (sc) IHF at millisecond resolution through single-molecule FRET (smFRET). Our findings reveal that the nucleoprotein complex exists in a ‘Slacked-Dynamic’ state throughout the observation window where many of them have switched between multiple ‘Wobbling States’ in the course of attainment of packaged form. A range of DNA ‘Flexure Angles’ has been calculated that give us vital insights regarding the nucleoid organization and transcriptional regulation in prokaryotes. -

Cell Cycle Regulation by the Bacterial Nucleoid
Available online at www.sciencedirect.com ScienceDirect Cell cycle regulation by the bacterial nucleoid David William Adams, Ling Juan Wu and Jeff Errington Division site selection presents a fundamental challenge to all terminate in the terminus region (Ter; 1808). Once organisms. Bacterial cells are small and the chromosome chromosome replication and segregation are complete (nucleoid) often fills most of the cell volume. Thus, in order to the cell is ready to divide. In Bacteria this normally occurs maximise fitness and avoid damaging the genetic material, cell by binary fission and in almost all species this is initiated division must be tightly co-ordinated with chromosome by the assembly of the tubulin homologue FtsZ into a replication and segregation. To achieve this, bacteria employ a ring-like structure (‘Z-ring’) at the nascent division site number of different mechanisms to regulate division site (Figure 1) [1]. The Z-ring then functions as a dynamic selection. One such mechanism, termed nucleoid occlusion, platform for assembly of the division machinery [2,3]. Its allows the nucleoid to protect itself by acting as a template for central role in division also allows FtsZ to serve as a nucleoid occlusion factors, which prevent Z-ring assembly over regulatory hub for the majority of regulatory proteins the DNA. These factors are sequence-specific DNA-binding identified to date [2,4]. Nevertheless, the precise ultra- proteins that exploit the precise organisation of the nucleoid, structure of the Z-ring and whether or not it plays a direct allowing them to act as both spatial and temporal regulators of role in force-generation during division remains contro- bacterial cell division. -

Analysis of a Multicomponent Thermostable DNA Polymerase III Replicase from an Extreme Thermophile*
THE JOURNAL OF BIOLOGICAL CHEMISTRY Vol. 277, No. 19, Issue of May 10, pp. 17334–17348, 2002 © 2002 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in U.S.A. Analysis of a Multicomponent Thermostable DNA Polymerase III Replicase from an Extreme Thermophile* Received for publication, October 23, 2001, and in revised form, February 18, 2002 Published, JBC Papers in Press, February 21, 2002, DOI 10.1074/jbc.M110198200 Irina Bruck‡, Alexander Yuzhakov§¶, Olga Yurieva§, David Jeruzalmi§, Maija Skangalis‡§, John Kuriyan‡§, and Mike O’Donnell‡§ʈ From §The Rockefeller University and ‡Howard Hughes Medical Institute, New York, New York 10021 This report takes a proteomic/genomic approach to polymerase III (pol III) structure and function has been ob- characterize the DNA polymerase III replication appa- tained from studies of the Escherichia coli replicase, DNA ratus of the extreme thermophile, Aquifex aeolicus. polymerase III holoenzyme (reviewed in Ref. 6). Therefore, a Genes (dnaX, holA, and holB) encoding the subunits re- brief overview of its structure and function is instructive for the ␦ ␦ quired for clamp loading activity ( , , and ) were iden- comparisons to be made in this report. In E. coli, the catalytic Downloaded from tified. The dnaX gene produces only the full-length subunit of DNA polymerase III is the ␣ subunit (129.9 kDa) product, , and therefore differs from Escherichia coli encoded by dnaE; it lacks a proofreading exonuclease (7). The dnaX that produces two proteins (␥ and ). Nonetheless, Ј Ј ⑀ ␦␦ proofreading 3 –5 -exonuclease activity is contained in the the A. aeolicus proteins form a complex. The dnaN ␣ ,␦␦ (27.5 kDa) subunit (dnaQ) that forms a 1:1 complex with (8  gene encoding the clamp was identified, and the ␣Ϫ⑀  9). -

Catalytic Domain of Plasmid Pad1 Relaxase Trax Defines a Group Of
Catalytic domain of plasmid pAD1 relaxase TraX defines a group of relaxases related to restriction endonucleases María Victoria Franciaa,1, Don B. Clewellb,c, Fernando de la Cruzd, and Gabriel Moncaliánd aServicio de Microbiología, Hospital Universitario Marqués de Valdecilla e Instituto de Formación e Investigación Marqués de Valdecilla, Santander 39008, Spain; bDepartment of Biologic and Materials Sciences, University of Michigan School of Dentistry, Ann Arbor, MI 48109; cDepartment of Microbiology and Immunology, University of Michigan Medical School, Ann Arbor, MI 48109; and dDepartamento de Biología Molecular e Instituto de Biomedicina y Biotecnología de Cantabria, Universidad de Cantabria–Consejo Superior de Investigaciones Científicas–Sociedad para el Desarrollo Regional de Cantabria, Santander 39011, Spain Edited by Roy Curtiss III, Arizona State University, Tempe, AZ, and approved July 9, 2013 (received for review May 30, 2013) Plasmid pAD1 is a 60-kb conjugative element commonly found in known relaxases show two characteristic sequence motifs, motif I clinical isolates of Enterococcus faecalis. The relaxase TraX and the containing the catalytic Tyr residue (which covalently attaches to primary origin of transfer oriT2 are located close to each other and the 5′ end of the cleaved DNA) and motif III with the His-triad have been shown to be essential for conjugation. The oriT2 site essential for relaxase activity (facilitating the cleavage reaction contains a large inverted repeat (where the nic site is located) by activation of the catalytic Tyr). This His-triad has been used as adjacent to a series of short direct repeats. TraX does not show a relaxase diagnostic signature (12). fi any of the typical relaxase sequence motifs but is the prototype of Plasmid pAD1 relaxase, TraX, was identi ed (9) and shown to oriT2 a unique family of relaxases (MOB ). -

Relaxation Complexes of Plasmids Colel and Cole2:Unique Site of The
Proc. Nat. Acad. Sci. USA Vol. 71, No. 10, pp. 3854-3857, October 1974 Relaxation Complexes of Plasmids ColEl and ColE2: Unique Site of the Nick in the Open Circular DNA of the Relaxed Complexes (Escherichia coli/supercoiled DNA/endonuclease/restriction enzyme/DNA replication) MICHAEL A. LOVETT, DONALD G. GUINEY, AND DONALD R. HELINSKI Department of Biology, University of California, San Diego, La Jolla, Calif. 92037 Communicated by Stanley L. Miller, June 26, 1974 ABSTRACT The product of the induced relaxation of volved in the production of a single-strand cleavage in the supercoiled DNA-protein relaxation complexes of colicino- initiation of replication and/or conjugal transfer of plasmid genic factors El (ColEl) and E2 (CoIE2) is an open circular DNA molecule with a strand-specific nick. Cleavage of the DNA. An involvement in such processes would most likely open circular DNA of each relaxed complex with the EcoRI require that the relaxation event take place at a unique site restriction endonuclease demonstrates that the single- in the DNA molecule. In this report, evidence will be pre- strand break is at a unique position. The site of the single- sented from studies using the EcoRI restriction endonuclease strand break in the relaxed ColEl complex is approximately the same distance from the EcoRI cleavage site as the that the strand-specific relaxation event takes place at a origin of ColEl DNA replication. unique site on both the ColEl and CoIE2 plasmid molecules. The location of this site, at least in the case of the ColEl Many of extrachromosomal circular DNA elements (plas- plasmid, is approximately the same distance from the single mids) of Escherichia coli, differing in molecular weight, num- EcoRI site as the origin of replication for this plasmid. -

DNA POLYMERASE III HOLOENZYME: Structure and Function of a Chromosomal Replicating Machine
Annu. Rev. Biochem. 1995.64:171-200 Copyright Ii) 1995 byAnnual Reviews Inc. All rights reserved DNA POLYMERASE III HOLOENZYME: Structure and Function of a Chromosomal Replicating Machine Zvi Kelman and Mike O'Donnell} Microbiology Department and Hearst Research Foundation. Cornell University Medical College. 1300York Avenue. New York. NY }0021 KEY WORDS: DNA replication. multis ubuni t complexes. protein-DNA interaction. DNA-de penden t ATPase . DNA sliding clamps CONTENTS INTRODUCTION........................................................ 172 THE HOLO EN ZYM E PARTICL E. .......................................... 173 THE CORE POLYMERASE ............................................... 175 THE � DNA SLIDING CLAM P............... ... ......... .................. 176 THE yC OMPLEX MATCHMAKER......................................... 179 Role of ATP . .... .............. ...... ......... ..... ............ ... 179 Interaction of y Complex with SSB Protein .................. ............... 181 Meclwnism of the yComplex Clamp Loader ................................ 181 Access provided by Rockefeller University on 08/07/15. For personal use only. THE 't SUBUNIT . .. .. .. .. .. .. .. .. .. .. .. .. .. .. .. .. .. .. .. .. .. .. .. 182 Annu. Rev. Biochem. 1995.64:171-200. Downloaded from www.annualreviews.org AS YMMETRIC STRUC TURE OF HOLO EN ZYM E . 182 DNA PO LYM ER AS E III HOLO ENZ YME AS A REPLIC ATING MACHINE ....... 186 Exclwnge of � from yComplex to Core .................................... 186 Cycling of Holoenzyme on the LaggingStrand -

Distinct Co-Evolution Patterns of Genes Associated to DNA Polymerase III Dnae and Polc Stefan Engelen1,2, David Vallenet2, Claudine Médigue2 and Antoine Danchin1,3*
Engelen et al. BMC Genomics 2012, 13:69 http://www.biomedcentral.com/1471-2164/13/69 RESEARCHARTICLE Open Access Distinct co-evolution patterns of genes associated to DNA polymerase III DnaE and PolC Stefan Engelen1,2, David Vallenet2, Claudine Médigue2 and Antoine Danchin1,3* Abstract Background: Bacterial genomes displaying a strong bias between the leading and the lagging strand of DNA replication encode two DNA polymerases III, DnaE and PolC, rather than a single one. Replication is a highly unsymmetrical process, and the presence of two polymerases is therefore not unexpected. Using comparative genomics, we explored whether other processes have evolved in parallel with each polymerase. Results: Extending previous in silico heuristics for the analysis of gene co-evolution, we analyzed the function of genes clustering with dnaE and polC. Clusters were highly informative. DnaE co-evolves with the ribosome, the transcription machinery, the core of intermediary metabolism enzymes. It is also connected to the energy-saving enzyme necessary for RNA degradation, polynucleotide phosphorylase. Most of the proteins of this co-evolving set belong to the persistent set in bacterial proteomes, that is fairly ubiquitously distributed. In contrast, PolC co- evolves with RNA degradation enzymes that are present only in the A+T-rich Firmicutes clade, suggesting at least two origins for the degradosome. Conclusion: DNA replication involves two machineries, DnaE and PolC. DnaE co-evolves with the core functions of bacterial life. In contrast PolC co-evolves with a set of RNA degradation enzymes that does not derive from the degradosome identified in gamma-Proteobacteria. This suggests that at least two independent RNA degradation pathways existed in the progenote community at the end of the RNA genome world. -

Glycolytic Pyruvate Kinase Moonlighting Activities in DNA Replication
Glycolytic pyruvate kinase moonlighting activities in DNA replication initiation and elongation Steff Horemans, Matthaios Pitoulias, Alexandria Holland, Panos Soultanas, Laurent Janniere To cite this version: Steff Horemans, Matthaios Pitoulias, Alexandria Holland, Panos Soultanas, Laurent Janniere. Gly- colytic pyruvate kinase moonlighting activities in DNA replication initiation and elongation. 2020. hal-02992157 HAL Id: hal-02992157 https://hal.archives-ouvertes.fr/hal-02992157 Preprint submitted on 10 Dec 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Glycolytic pyruvate kinase moonlighting activities in DNA replication initiation and elongation Steff Horemans1, Matthaios Pitoulias2, Alexandria Holland2, Panos Soultanas2¶ and Laurent Janniere1¶ 1 : Génomique Métabolique, Genoscope, Institut François Jacob, CEA, CNRS, Univ Evry, Université Paris-Saclay, 91057 Evry, France 2 : Biodiscovery Institute, School of Chemistry, University of Nottingham, University Park, Nottingham NG7 2RD, UK Short title: PykA moonlighting activity in DNA replication Key Words: DNA replication; replication control; central carbon metabolism; glycolytic enzymes; replication enzymes; cell cycle; allosteric regulation. ¶ : Corresponding authors Laurent Janniere: [email protected] Panos Soultanas : [email protected] 1 SUMMARY Cells have evolved a metabolic control of DNA replication to respond to a wide range of nutritional conditions. -
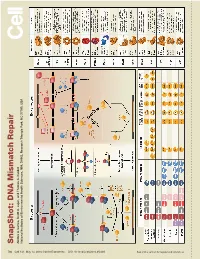
S Na P S H O T: D N a Mism a Tc H R E P a Ir
SnapShot: Repair DNA Mismatch Scott A. Lujan, and Thomas Kunkel A. Larrea, Andres Park, NC 27709, USA Triangle Health Sciences, NIH, DHHS, Research National Institutes of Environmental 730 Cell 141, May 14, 2010 ©2010 Elsevier Inc. DOI 10.1016/j.cell.2010.05.002 See online version for legend and references. SnapShot: DNA Mismatch Repair Andres A. Larrea, Scott A. Lujan, and Thomas A. Kunkel National Institutes of Environmental Health Sciences, NIH, DHHS, Research Triangle Park, NC 27709, USA Mismatch Repair in Bacteria and Eukaryotes Mismatch repair in the bacterium Escherichia coli is initiated when a homodimer of MutS binds as an asymmetric clamp to DNA containing a variety of base-base and insertion-deletion mismatches. The MutL homodimer then couples MutS recognition to the signal that distinguishes between the template and nascent DNA strands. In E. coli, the lack of adenine methylation, catalyzed by the DNA adenine methyltransferase (Dam) in newly synthesized GATC sequences, allows E. coli MutH to cleave the nascent strand. The resulting nick is used for mismatch removal involving the UvrD helicase, single-strand DNA-binding protein (SSB), and excision by single-stranded DNA exonucleases from either direction, depending upon the polarity of the nick relative to the mismatch. DNA polymerase III correctly resynthesizes DNA and ligase completes repair. In bacteria lacking Dam/MutH, as in eukaryotes, the signal for strand discrimination is uncertain but may be the DNA ends associated with replication forks. In these bacteria, MutL harbors a nick-dependent endonuclease that creates a nick that can be used for mismatch excision. Eukaryotic mismatch repair is similar, although it involves several dif- ferent MutS and MutL homologs: MutSα (MSH2/MSH6) recognizes single base-base mismatches and 1–2 base insertion/deletions; MutSβ (MSH2/MSH3) recognizes insertion/ deletion mismatches containing two or more extra bases. -

Effects of Abasic Sites and DNA Single-Strand Breaks on Prokaryotic RNA Polymerases (Apurinic/Apyrimidinic Sites/Transcription/DNA Damage) WEI ZHOU* and PAUL W
Proc. Natl. Acad. Sci. USA Vol. 90, pp. 6601-6605, July 1993 Biochemistry Effects of abasic sites and DNA single-strand breaks on prokaryotic RNA polymerases (apurinic/apyrimidinic sites/transcription/DNA damage) WEI ZHOU* AND PAUL W. DOETSCH*tt Departments of *Biochemistry and tRadiation Oncology and tDivision of Cancer Biology, Emory University School of Medicine, Atlanta, GA 30322 Communicated by Philip C. Hanawalt, April 19, 1993 ABSTRACT Abasic sites are thought to be the most fre- effects of such damage on both transcription initiation and quently occurring cellular DNA damage and are generated elongation processes (19, 20). We wished to determine the spontaneously or as the result of chemical or radiation damage effects of AP sites on the transcription elongation process by to DNA. In contrast to the wealth of information that exists on using a defined system that would provide direct information the effects of abasic sites on DNA polymerases, very little is regarding the ability of RNA polymerases to bypass or be known about how these lesions interact with RNA polymerases. blocked by AP sites and, if bypass was observed, to deter- An in vitro transcription system was used to determine the mine the nature of the base insertion opposite such lesions. effects of abasic sites and single-strand breaks on transcrip- For these studies, we constructed a DNA template containing tional elongation. DNA templates were constructed containing a single AP site placed at a unique location downstream from single abasic sites or nicks placed at unique locations down- either the SP6 or tac promoter and carried out in vitro stream from two different promoters and were transcribed by transcription experiments with SP6 and E. -

Coordination Between Nucleotide Excision Repair And
RESEARCH ARTICLE Coordination between nucleotide excision repair and specialized polymerase DnaE2 action enables DNA damage survival in non-replicating bacteria Asha Mary Joseph1, Saheli Daw1, Ismath Sadhir1,2, Anjana Badrinarayanan1* 1National Centre for Biological Sciences - Tata Institute of Fundamental Research, Bangalore, India; 2Max Planck Institute for Terrestrial Microbiology, LOEWE Centre for Synthetic Microbiology (SYNMIKRO), Marburg, Germany Abstract Translesion synthesis (TLS) is a highly conserved mutagenic DNA lesion tolerance pathway, which employs specialized, low-fidelity DNA polymerases to synthesize across lesions. Current models suggest that activity of these polymerases is predominantly associated with ongoing replication, functioning either at or behind the replication fork. Here we provide evidence for DNA damage-dependent function of a specialized polymerase, DnaE2, in replication- independent conditions. We develop an assay to follow lesion repair in non-replicating Caulobacter and observe that components of the replication machinery localize on DNA in response to damage. These localizations persist in the absence of DnaE2 or if catalytic activity of this polymerase is mutated. Single-stranded DNA gaps for SSB binding and low-fidelity polymerase-mediated synthesis are generated by nucleotide excision repair (NER), as replisome components fail to localize in the absence of NER. This mechanism of gap-filling facilitates cell cycle restoration when cells are released into replication-permissive conditions. Thus, such cross-talk (between activity of NER and specialized polymerases in subsequent gap-filling) helps preserve genome integrity and enhances survival in a replication-independent manner. *For correspondence: [email protected] Introduction Competing interests: The DNA damage is a threat to genome integrity and can lead to perturbations to processes of replica- authors declare that no tion and transcription. -

Dnag Primase—A Target for the Development of Novel Antibacterial Agents
antibiotics Review DnaG Primase—A Target for the Development of Novel Antibacterial Agents Stefan Ilic, Shira Cohen, Meenakshi Singh, Benjamin Tam, Adi Dayan and Barak Akabayov * ID Department of Chemistry, Ben-Gurion University of the Negev, Beer-Sheva 8410501, Israel; [email protected] (S.I.); [email protected] (S.C.); [email protected] (M.S.); [email protected] (B.T.) [email protected] (A.D.) * Correspondence: [email protected]; Tel.: +972-8-647-2716 Received: 18 July 2018; Accepted: 9 August 2018; Published: 13 August 2018 Abstract: The bacterial primase—an essential component in the replisome—is a promising but underexploited target for novel antibiotic drugs. Bacterial primases have a markedly different structure than the human primase. Inhibition of primase activity is expected to selectively halt bacterial DNA replication. Evidence is growing that halting DNA replication has a bacteriocidal effect. Therefore, inhibitors of DNA primase could provide antibiotic agents. Compounds that inhibit bacterial DnaG primase have been developed using different approaches. In this paper, we provide an overview of the current literature on DNA primases as novel drug targets and the methods used to find their inhibitors. Although few inhibitors have been identified, there are still challenges to develop inhibitors that can efficiently halt DNA replication and may be applied in a clinical setting. Keywords: DNA replication; DnaG primase; bacterial inhibitors; antibacterial agents; antibiotics 1. Introduction The complex process of identifying antibacterial compounds begins with the selection of potential targets, which must be essential, selective over human homologues, susceptible to drugs, and have a low propensity to develop a rapid resistance [1].