Inocybe Praetervisa Group E a Clade of Four Closely Related Species with Partly Different Geographical Distribution Ranges in Europe
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Preface to Artificial Key to Common and Noteworthy Species of Inocybe from the Pacific Northwest
Preface to Artificial Key to Common and Noteworthy Species of Inocybe from the Pacific Northwest This key is aimed at an audience familiar with the determination of agarics in general but unfamiliar with Inocybe. The key stresses gross morphological characters as I think appropriate before yielding to taxa that are better distinguished microscopically. 43 species are enumerated below and several others are mentioned, but probably over 100 occur in the Pacific Northwest, a region circumscribed to include British Columbia, Washington, Idaho, western Montana, Oregon, and northern California. Of the 43 species in the key, few are endemic to the region based on gross morphological species concepts. However, the key is recommended for use with Pacific Northwest material. Many eastern North American species of Inocybe, for example, do not occur in the Pacific Northwest and are excluded from this treatment. The genus Inocybe (Fr.) Fr. traditionally has encompassed dull brown-spored agarics that are ectomycorrhizal and frequently occur on soil; exhibit a dry pileus that is often rimose, fibrillose, or scaly; and have a distinctive smell that is often spermatic or less often fruity, sweet, aromatic, like bruised Geranium leaves, like Lycoperdon, or green corn. Species of Hebeloma differ by their gelatinous pileus, often radish smell, typically verrucose basidiospores, and absence of metuloid cystidia. Decomposers such as Phaeomarasmius and Flammulaster differ by their occurrence on woody debris and lack of metuloid cystidia. The Crepidotaceae, including Pleuroflammula and Simocybe, is the closest related group to Inocybe, which I treat as a separate family in its own right (see Matheny et al. (2006) Mycologia 98:982- 995). -

Plateau Mountain Plant List
Plateau ALBERTA WILDERNESS Mountain – Plant list ASSOCIATION Plants of Plateau Mountain Ecological Reserve (Plateau Mountain Ecological Reserve Management Plan – Alberta Environment) Alpine Anemone Anemone drummondii Drummond's Rock Cress Alpine Arnica Arnica angustifolia Drummond's Rush Alpine Bistort Polygonum viviparum Dwarf Birch Betula glandulosa Alpine Blue Grass Poa alpina Dwarf Bitter root Alpine Everlasting Antennaria sp. Dwarf Hawk's Beard Alpine Fleabane Erigeron pallens Dwarf Sawwort Saussurea nuda Alpine Forget-me not Myosotis alpestris Dwarf saw-wort Saussurea nuda Alpine Goldenrod Solidago multiradianta Dwarf Scouring-rush Equisetum scirpoides Alpine Hawkweed Early Blue Grass Alpine Milk-vetch Astragalus alpinus Early Blue Violet Viola adunca Alpine mouse-eared chickweed Cerastium beeringianum Early Cinquefoil Potertilla concinna Alpine Speedwell Veronica alpina Elephant's head Pedicularis groenlandica Alpine speedwell Veronica alpina Elephant's-head Lousewort Pedicualaris groenlandica Alpine Timothy Elgelmann Spruce Picea engelmanii American Vetch Entire-leaved Groundsel Androsace Androsace chamaejasme Everlasting Antennaria luzuloides Arctic Aster False Dandelion Agoseris aurantiaca Arctic Blue Glass Felwort Gentianela amarella Arctic Butterweed Few-flowered Milk-vetch Astragalus sp. Balsam Groundsel Few-seeded Whitlow-grass Draba oligosperma Balsam Poplar Populus balsamifera Field Chickweed Stellaria sp. Barratt's Willow Fireweed Locoweed Oxytropis sp. Bearberry Arctostaphylos uva-ursi Flame-colored Lousewort Pedicularis -

Survey of Dryas Octopetala- Dominated Plant Communities in The
Botanica Pacifica. A journal of plant science and conservation. 2015. 4(1) DOI: 10.17581/bp.2015.04102 Survey of Dryas octopetala - dominated plant communities in the European and North-West-Siberian Arctic Natalia E. Koroleva Natalia E. Koroleva ABSTRACT email: [email protected] 18 associations of Dryas octopetala-dominated communities, which belong to 3 classes and 4 alliances, were observed in the European Arctic and in north- Polar-Alpine Botanical Garden-Institute, western Siberia. Syntaxonomical differentiation is linked with the ecogeographical Kirovsk, Russia and floristical division of the Arctic: alliance Kobresio-Dryadion of Carici rupestris– Kobresietea bellardii has its main distribution in European arctic and typical tundra and in the oroarctic belt of mountains, whilst the area of alliance Dryado Manuscript received: 25.08.2014 octopetalae–Caricion arctisibiricae lies in the southern and typical tundra and oroarctic Review completed: 03.02.2015 Accepted for publication: 12.02.2015 belt on the north of Pechora Plain, in theYugorskii Peninsula, Pay-Khoy Mts. and Published online: 20.04.2015 Polar Ural. Dryas octopetala-dominated communities lacking characteristic species of Carici rupestris–Kobresietea bellardii have been related to the alliance Cassiopo- Salicion herbaceae (Salicetea herbaceae) and alliance Loiseleurio-Diapension (Loseleurio- Vaccinietea). In the latter a suballiance Salicenion nummulariae has been proposed comprising xero-, achyonophytic communities on sandy and gravelly substrata in the southern and typical tundra of the north-east of European Russia. A synoptical table is provided. Keywords: Braun-Blanquet, Dryas octopetala-dominated communities, European Arctic, north-western Siberia, Carici rupestris–Kobresietea bellardii, Loseleurio–Vaccinietea, Salicetea herbaceae РЕЗЮМЕ Cообщества с доминированием Dryas octopetala в европейском секторе Арк ти ки и Субарктики и на севере Западной Сибири принадлежат к 18 ассоци а циям в 3 классах и 4 союзах. -

Final Environmental Impact Statement
Appendix M Biological Assessment BIOLOGICAL ASSESSMENT FOR PROJECT ICEBREAKER Lake Erie and City of Cleveland Cuyahoga County, Ohio TABLE OF CONTENTS TABLE OF CONTENTS .................................................................................................................... ii TABLES ............................................................................................................................................ iv FIGURES .......................................................................................................................................... iv COMMONLY USED ACRONYMS and ABBREVIATIONS ............................................................... v 1.0 INTRODUCTION ................................................................................................................................................... 1 1.1 PURPOSE OF THE BIOLOGICAL ASSESSMENT ................................................................ 1 1.2 PROPOSED ACTION .............................................................................................................. 2 1.2.1 Federal Action .................................................................................................................... 2 1.2.2 LEEDCo’s Action ............................................................................................................... 2 2.0 PROJECT DESCRIPTION .................................................................................................................................... 3 2.1 DESCRIPTION OF MAJOR -

Swing Through
Swing Through 20m Swing Through is an interactive agility garden that connects the user to Canada’s diverse landscape, as well as its major economic industry. The garden is a series of thirteen finished lumber posts that dangle from a large steel structure, creating “tree swings”. On the swings are climbing holds where visitors can use the holds to climb up and across the tree swings. Directly under the tree swings are thirteen colour-coordinated stumps that give the user an extra boost, if needed. The thirteen timber tree swings represent Canada’s ten provinces and three territories by using wood from the official provincial and territorial trees. Surrounding this structure of Canadian trees is a garden divided into thirteen sections displaying the native plants of each province and territory. This representative regional plantings encompassing the swings, creating a soft edge. 10m Swing Through allows visitors to touch, smell, and play with the various YT NT NU BC AB SK MB ON QC NL NB PE NS natural elements that make our country so green, prosperous and beautiful. PLAN | 1:75 Yukon Nunavut Alberta Manitoba Quebec New Brunswick Nova Scotia Tree: Subapline fir, Abies lasiocarpa Tree: Balsam Poplar, Populus balsamifera Tree: Lodgepole pine, Pinus contorta Tree: Balsam fir, Abies balsamea Tree: Yellow birch, Betula alleghaniensis Tree: Balsam fir, Abies balsamea Tree: Red spruce, Picea rubens Plants: Epilobium angustifolium, Plants: Saxifraga oppositifolia, Rubus Plants: Rosa acicularis Prunus virginiana, Plants: Pulsatilla ludoviciana, -

Herbariet Publ 2010-2019 (PDF)
Publikationer 2019 Amorim, B. S., Vasconcelos, T. N., Souza, G., Alves, M., Antonelli, A., & Lucas, E. (2019). Advanced understanding of phylogenetic relationships, morphological evolution and biogeographic history of the mega-diverse plant genus Myrcia and its relatives (Myrtaceae: Myrteae). Molecular phylogenetics and evolution, 138, 65-88. Anderson, C. (2019). Hiraea costaricensis and H. polyantha, Two New Species Of Malpighiaceae, and circumscription of H. quapara and H. smilacina. Edinburgh Journal of Botany, 1-16. Athanasiadis, A. (2019). Carlskottsbergia antarctica (Hooker fil. & Harv.) gen. & comb. nov., with a re-assessment of Synarthrophyton (Mesophyllaceae, Corallinales, Rhodophyta). Nova Hedwigia, 108(3-4), 291-320. Athanasiadis, A. (2019). Amphithallia, a genus with four-celled carpogonial branches and connecting filaments in the Corallinales (Rhodophyta). Marine Biology Research, 15(1), 13-25. Bandini, D., Oertel, B., Moreau, P. A., Thines, M., & Ploch, S. (2019). Three new hygrophilous species of Inocybe, subgenus Inocybe. Mycological Progress, 18(9), 1101-1119. Baranow, P., & Kolanowska, M. (2019, October). Sertifera hirtziana (Orchidaceae, Sobralieae), a new species from southeastern Ecuador. In Annales Botanici Fennici (Vol. 56, No. 4-6, pp. 205-209). Barboza, G. E., García, C. C., González, S. L., Scaldaferro, M., & Reyes, X. (2019). Four new species of Capsicum (Solanaceae) from the tropical Andes and an update on the phylogeny of the genus. PloS one, 14(1), e0209792. Barrett, C. F., McKain, M. R., Sinn, B. T., Ge, X. J., Zhang, Y., Antonelli, A., & Bacon, C. D. (2019). Ancient polyploidy and genome evolution in palms. Genome biology and evolution, 11(5), 1501-1511. Bernal, R., Bacon, C. D., Balslev, H., Hoorn, C., Bourlat, S. -
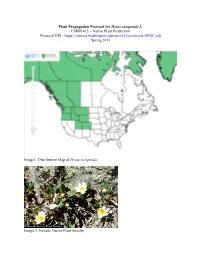
Plant Propagation Protocol for Dryas Octopetala L. ESRM 412 – Native Plant Production Protocol URL
Plant Propagation Protocol for Dryas octopetala L. ESRM 412 – Native Plant Production Protocol URL: https://courses.washington.edu/esrm412/protocols/DROC.pdf Spring 2015 Image1. Distribution Map of Dryas octopetala Image 2. Nevada Native Plant Society TAXONOMY Plant Family [3] Scientific Name Rosaceae Common Name Rose Family Species Scientific Name Scientific Name Dryas octopetala L. [3] Varieties * Dryas octopetala var. angustifolia C.L. Hitchc. [3] Dryas octopetala var. hookeriana (Juz) Hulten Sub-species Dryas octopetala f. argentea (Blytt) Hulten * Dryas octopetala subsp. alaskensis (Porsild) Hulten [3] Cultivar Common Synonym(s) Dradetum octopetalae Keiner Common Name(s) White mountain-avens, Eightpetal mountain-avens, Mountain dryas Species Code (as per USDA Plants DROC database) GENERAL INFORMATION Geographical range Alaska, Washington, Oregon, Colorado Alpine regions in the Pacific Northwest N. Cascades, and Rocky Mountain ranges Ecological distribution Mid-montane to Alpine zone Climate and elevation range Elevation: 3,500 m [6] 100 m and less Climate: sites with low snow cover on calcareous or basic soils [6] Local habitat and abundance Full sun Dry, well-drained, sandy or gravelly soils Spreads rapidly [6] Dominant or co-dominant species within its range [6] Associated species: Dwarf willow Plant strategy type / successional Nitrogen fixer: forms association with Frankia [5] stage Colonizer of barren slopes at high elevations [5] Plant characteristics Forb/herb, Shrub, Subshrub Perrennial Forms mats up to 3 ft. wide and 8 in. tall. [1] 1 cream or white flower at the end of each 2-8 inch leafless flower stalk. [2] Flowers bloom June-July Summer fruits fluffy and feathery Seeds are wind-dispersed [7] Leaves are oval-shaped, leathery with rounded teeth and a white underside. -

Improving Phylogenetic Inference of Mushrooms with RPB1 and RPB2 Nucleotide Sequences (Inocybe; Agaricales)
Molecular Phylogenetics and Evolution 35 (2005) 1–20 www.elsevier.com/locate/ympev Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe; Agaricales) P. Brandon Matheny¤,1 Biology Department, Box 351330, University of Washington, Seattle, WA 98195-5325, USA Received 9 July 2003; revised 15 May 2004 Available online 18 January 2005 Abstract Approximately 3000 bp across 84 taxa have been analyzed for variable regions of RPB1, RPB2, and nLSU-rDNA to infer phylo- genetic relationships in the large ectomycorrhizal mushroom genus Inocybe (Agaricales; Basidiomycota). This study represents the Wrst eVort to combine variable regions of RPB1 and RPB2 with nLSU-rDNA for low-level phylogenetic studies in mushroom-form- ing fungi. Combination of the three loci increases non-parametric bootstrap support, Bayesian posterior probabilities, and resolution for numerous clades compared to separate gene analyses. These data suggest the evolution of at least Wve major lineages in Inocybe— the Inocybe clade, the Mallocybe clade, the Auritella clade, the Inosperma clade, and the Pseudosperma clade. Additionally, many clades nested within each major lineage are strongly supported. These results also suggest the family Crepiodataceae sensu stricto is sister to Inocybe. Recognition of Inocybe at the family level, the Inocybaceae, is recommended. 2004 Elsevier Inc. All rights reserved. Keywords: Cortinariaceae; Fungi; Inocybaceae; nLSU-rDNA; RNA polymerase II; Systematics 1. Introduction room taxa in Inocybe and outgroups of the Agaricales, or euagarics clade, has been extended to include partial Nuclear genes that encode the two largest subunits of sequences of RPB1, RPB2, and nuclear large subunit RNA polymerase II are proving useful to infer the phy- ribosomal DNA (nLSU). -

Prächtiger Frauenschuh 7-18 © Verein Zum Schutz Der Bergwelt E.V
ZOBODAT - www.zobodat.at Zoologisch-Botanische Datenbank/Zoological-Botanical Database Digitale Literatur/Digital Literature Zeitschrift/Journal: Jahrbuch des Vereins zum Schutze der Alpenpflanzen und -Tiere Jahr/Year: 1972 Band/Volume: 37_1972 Autor(en)/Author(s): Eberle Georg Artikel/Article: Prächtiger Frauenschuh 7-18 © Verein zum Schutz der Bergwelt e.V. download unter www.vzsb.de/publikationen.php und www.zobodat.at Prächtiger Frauenschuh (Cypripedium calceolus L.) Von Georg Eberle, Wetzlar Inhalt I. Sonderstellung der Frauenschuhe (Cypripedioideae) unter den Orchideen und ihre Gliederung II. Areale der Frauenschuhe (Cypripedioideae) III. Areal und Standorte von Cypripedium calceolus L. IV. Der blühende Frauenschuh und seine Vergesellschaftung V. Kesselfallenblume Frauenschuh: Gestalt und Funktion VI. Frucht, Same und Jugendentwicklung des Frauenschuhs VII. Gefährdung und Schutz des Frauenschuhs VIII. Schrifttum ie oft bin ich ihr wohl in meinem Leben begegnet, der Wunderblume Frauenschuh? W Und jedesmal war es ein neues großes Ereignis - und mit jeder künfligen Begeg nung wird es nicht anders sein. An ihm, dem tiefdringenden Erlebnis, sind nicht nur das Schauen der Formen und Farben sowie das Wahrnehmen des Duftes beteiligt, sondern auch das Wissen um das Leben dieses Wesens, seines schicksalhaflen Verwobenseins in seine Verwandtschafl und seine Umwelt, zu der auch - der Mensch gehört. I. Sonderstellung der Frauenschuhe (Cypripedioideae) unter den Orchideen und ihre Gliederung Unter den etwa 65 in Mittel- und Nordeuropa vorkommenden -

Dryas Octopetala Subsp. Alaskensis: Potential for a Hardy, Novel Flower
Dryas octopetala subsp. alaskensis: Potential for a Hardy, Novel Flower Eamon Spanier-Scanlon, Student of Plant Science University of Minnesota, St. Paul, MN 5/13/2019 Executive Summary Dryas octopetala, Mountain Avens, is a new sprawling variety of ground cover which may become available in northern climates near you. This extremely cold-tolerant plant will spread easily in your garden or within a larger pot with other plants. With petite white flowers blooming late in the spring, and placed gingerly upon thin red stalks, their beauty persists throughout the summer. This perennial ground cover will return year after year, regardless of the harshest winters. Purchase a few and watch as they creep around your yard, or simply buy one and propagate your own, with ease, through cuttings. The northern consumer may often feel left out by the lack of perennial flowering plants, but with this new species you can soon become the envy of your friends residing in the south. Minimal input is required for these beautiful white flowers to flourish in your domain. Pick up your own in a local floral retailer and fall in love over and over, each spring, as they bound back to life. I. Introduction A. Species The distribution of the genus Dryas is worldwide; they are often found in harsh areas such as arctic and tundra regions (Springer et al., 2019). Dryas octopetala is currently on the market as a decorative, flowering plant, although subsp. alaskensis is not. It can be purchased as a seed from multiple vendors, coming in such names as “Mountain Evans,” available through GeoSeed company (https://www.geoseed.com). -

BULLETIN of the AMERICAN ROCK GARDEN SOCIETY
. BULLETIN of the AMERICAN ROCK GARDEN SOCIETY VOL. 25 October, 1967 No. 4 FALL COLOR IN THE ROCK GARDEN—Anna R. Zollingei 109 DRYAS—H. Lincoln Foster 110 FLORA OF THE KRKONOSE NATIONAL PARK Ing. Frantisek Prochazka 113 SCABIOSAS FOR THE ROCK GARDEN—Robert M. Senior 115 TRULY A ROCK GARDEN—A. M. S 117 BOOK REVIEW—Dorothy Metheny 124 OUR TINIEST EASTERN MOUNTAIN VIOLET Edgar T. Wherry 125 LETTERS 126 EASTERN GARDENS VISITED—Grace M. Conboy 128 A POSTSCRIPT—C. R. Worth 134 JERVIS, A HEMLOCK OF ODD FORM—G. G. Nearing 135 INTERCHANGE 136 MORE ABOUT WEST AUSTRALIAN PLANTS Brian Halliwell 137 NOTES FROM THE NORTHWEST—Elizabeth Peterson 138 OMNIUM-GATHERUM 140 DIRECTORATE BULLETIN Editor Emeritus DR. EDGAR T. WHERRY, University of Pennsylvania, Philadelphia 4, Pa. Editor ALBERT M. SUTTON 9608 26th Ave. N. W., Seattle, Washington 98107 AMERICAN ROCK GARDEN SOCIETY President Emeritus HAROLD EPSTEIN, 5 Forest Court, Larchmont, New York President H. LINCOLN FOSTER, Falls Village, Conn. 06031 Secretary RICHARD W. REDFIELD, BOX 26, Closter, N. J. 07624 Treasurer ALEX D. REID, 260 Boulevard, Mountain Lakes, N. J. Vice-Presidents MRS. HARRY HAYWARD F. O. PEARCE BRIAN O. MULLIGAN RICHARD LANGFELDER MRS. LEE ARMIGER Directors Terra Expires 1968 BURR B. BRONSON BERNARD HARKNESS LEE H. RADEN Term Expires 1969 MRS. SALLIE D. ALLEN JEROME A. LUKINS HENRY R. FULLER Term Expires 1970 JOHN P. OSBORNE WALTER A. KOLAGA CLAUDE A. BARR Director of Seed Exchange LAWRENCE CROCKER 3355 Jacksonville Hy., Medford, Oregon 97501 Director of Slide Collection MRS. RONALD S. GRAY 64 North Street, North Reading, Massachusetts 01864 REGIONAL CHAIRMEN Northwestern I JOSEPH A. -

List of Plants for Great Sand Dunes National Park and Preserve
Great Sand Dunes National Park and Preserve Plant Checklist DRAFT as of 29 November 2005 FERNS AND FERN ALLIES Equisetaceae (Horsetail Family) Vascular Plant Equisetales Equisetaceae Equisetum arvense Present in Park Rare Native Field horsetail Vascular Plant Equisetales Equisetaceae Equisetum laevigatum Present in Park Unknown Native Scouring-rush Polypodiaceae (Fern Family) Vascular Plant Polypodiales Dryopteridaceae Cystopteris fragilis Present in Park Uncommon Native Brittle bladderfern Vascular Plant Polypodiales Dryopteridaceae Woodsia oregana Present in Park Uncommon Native Oregon woodsia Pteridaceae (Maidenhair Fern Family) Vascular Plant Polypodiales Pteridaceae Argyrochosma fendleri Present in Park Unknown Native Zigzag fern Vascular Plant Polypodiales Pteridaceae Cheilanthes feei Present in Park Uncommon Native Slender lip fern Vascular Plant Polypodiales Pteridaceae Cryptogramma acrostichoides Present in Park Unknown Native American rockbrake Selaginellaceae (Spikemoss Family) Vascular Plant Selaginellales Selaginellaceae Selaginella densa Present in Park Rare Native Lesser spikemoss Vascular Plant Selaginellales Selaginellaceae Selaginella weatherbiana Present in Park Unknown Native Weatherby's clubmoss CONIFERS Cupressaceae (Cypress family) Vascular Plant Pinales Cupressaceae Juniperus scopulorum Present in Park Unknown Native Rocky Mountain juniper Pinaceae (Pine Family) Vascular Plant Pinales Pinaceae Abies concolor var. concolor Present in Park Rare Native White fir Vascular Plant Pinales Pinaceae Abies lasiocarpa Present