Downloads-Blood- Pressure.Html
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Global Comparison of Non-Sovereign Island Territories: the Search for ‘True Equality’
Island Studies Journal, 15(1), 2020, 43-66 A global comparison of non-sovereign island territories: the search for ‘true equality’ Malcom Ferdinand CNRS, Paris, France [email protected] Gert Oostindie KITLV, the Netherlands Leiden University, the Netherlands [email protected] (corresponding author) Wouter Veenendaal KITLV, the Netherlands Leiden University, the Netherlands [email protected] Abstract: For a great majority of former colonies, the outcome of decolonization was independence. Yet scattered across the globe, remnants of former colonial empires are still non-sovereign as part of larger metropolitan states. There is little drive for independence in these territories, virtually all of which are small island nations, also known as sub-national island jurisdictions (SNIJs). Why do so many former colonial territories choose to remain non-sovereign? In this paper we attempt to answer this question by conducting a global comparative study of non-sovereign jurisdictions. We start off by analyzing their present economic, social and political conditions, after which we assess local levels of (dis)content with the contemporary political status, and their articulation in postcolonial politics. We find that levels of discontent and frustration covary with the particular demographic, socio- economic and historical-cultural conditions of individual territories. While significant independence movements can be observed in only two or three jurisdictions, in virtually all cases there is profound dissatisfaction and frustration with the contemporary non-sovereign arrangement and its outcomes. Instead of achieving independence, the territories’ real struggle nowadays is for obtaining ‘true equality’ with the metropolis, as well as recognition of their distinct cultural identities. -

Irma Omerasevic Bosnia and Herzegovina Bosnia And
Irma Omerasevic Bosnia and Herzegovina Bosnia and Herzegovina is located in Southeastern Europe near Croatia and Italy. The terrain consists of mountains, valleys, agricultural land, forests, and rivers. Although Bosnia and Herzegovina has many different types of lands, most of Bosnia and Herzegovina is covered with agricultural land, as it houses animals that are used for consumption and selling purposes. Most of the citizens in Bosnia and Herzegovina speak in the Bosnian language. There are many countries surrounding Bosnia and Herzegovina that speak a similar language. These countries include Croatia, Serbia, and Slovenia. Since they are so similar, the languages of these countries can also be spoken in Bosnia and Herzegovina alongside the Bosnian language. Bosnia and Herzegovina is projected to have a population of 3.8 million people by July of 2020, according to the Central Intelligence Agency. Of the population, most of the urban population resides in the valleys and in wide areas around agriculture, as there is more access to water. Most of the rural population tends to reside in the mountainous and forest regions. While few parts of the country are still dealing with the effects of poverty after the Bosnian War in the 1990’s, urban populations have more of an advantage. Urban populations reside around agricultural land and valleys, which happen to have more towns and civilians. This land is also more developed than the mountainous regions. The valley has more access to water because there are plenty of rivers nearby. Even though water is accessible, the country still implements archaic ways of life when it comes to the water supply. -

The KOF Education System Factbook: Bosnia and Herzegovina
The KOF Education System Factbook: Bosnia and Herzegovina Edition 1, December 2019 ETH Zurich KOF Swiss Economic Institute LEE G 116 Leonhardstrasse 21 8092 Zurich, Switzerland Phone +41 44 632 42 39 Fax +41 44 632 12 18 www.kof.ethz.ch [email protected] Table of Contents FOREWORD .................................................................................................................... IV EDITING AND ACKNOWLEDGEMENTS .......................................................................... V 1. The Bosnia and Herzegovina Economy and its Political System ............................. 1 1.1 The Bosnia and Herzegovina Economy ................................................................... 1 1.2 The Labour Market .................................................................................................. 3 1.2.1 Overview of Bosnia and Herzegovina’s Labour Market .................................... 4 1.2.2 The Youth Labour Market ................................................................................. 6 1.2.3 The KOF Youth Labour Market Index (KOF YLMI) for Bosnia and Herzegovina 7 1.3 The Political System ................................................................................................ 9 1.3.1 Overview of the Bosnia and Herzegovina Political System ............................... 9 1.3.2 Politics and Goals of the Education System ....................................................11 2. Formal System of Education ......................................................................................12 -

Democracy in the Cities: a New Proposal for Chinese Reform
Bloch and TerBush: Democracy in the Cities: A New Proposal for Chinese Reform DEMOCRACY IN THE CITIES: A NEW PROPOSAL FOR CHINESE REFORM DAVID S. BLOCH* THOMAS TERBUSH** [I]t has been no easy job for a big developing country like China with a population of nearly 1.3 billion to have so considerably improved its hu- man rights situation in such a short period of time. -President Hu Jintao, People's Republic of China.' I. THE DILEMMA OF CHINESE DEMOCRACY A great deal has been written on the question of Chinese democracy. In practice and theory, democracy in China is enormously significant.2 This is because China is a rising military threat whose interests are often counter to those of the United States, as well as a demographic powerhouse with as much as a quarter of the world's population. In Mainland China, "the current official mythology.., holds that Chi- nese culture and democracy are incompatible."3 Many Mainland Chinese apparently believe that China's Confucian traditions are inconsistent with democratic practices--an idea with a pedigree that traces both to China's Attorney, Gray Cary Ware & Freidenrich LLP, Palo Alto, California; admitted in Cali- fornia and the District of Columbia; B.A., Reed College (41BK); M.P.H., J.D. with honors, The George Washington University; 1997 Fellow in International Trade Law, University In- stitute of European Studies, Turin, Italy. ** Economist and Senior Analyst, Electric Power Research Institute, Palo Alto, California and Tokyo, Japan; M.A., Ph.D., George Mason University. 1. President Hu Jintao, Enhanced Mutual Understanding and Trust Towards a Conserva- tive and Cooperative Relationship Between China and the United States, translated at www.asiasociety.org/speeches/jintao.htm (speech given by then-Vice President Hu Jintao). -

Curating the CIA World Factbook 29 the International Journal of Digital Curation Issue 3, Volume 4 | 2009
Curating the CIA World Factbook 29 The International Journal of Digital Curation Issue 3, Volume 4 | 2009 Curating the CIA World Factbook Peter Buneman, Heiko Müller, School of Informatics, University of Edinburgh Chris Rusbridge, Digital Curation Centre, University of Edinburgh Abstract The CIA World Factbook is a prime example of a curated database – a database that is constructed and maintained with a great deal of human effort in collecting, verifying, and annotating data. Preservation of old versions of the Factbook is important for verification of citations; it is also essential for anyone interested in the history of the data such as demographic change. Although the Factbook has been published, both physically and electronically, only for the past 30 years, we appear in danger of losing this history. This paper investigates the issues involved in capturing the history of an evolving database and its application to the CIA World Factbook. In particular it shows that there is substantial added value to be gained by preserving databases in such a way that questions about the change in data, (longitudinal queries) can be readily answered. Within this paper, we describe techniques for recording change in a curated database and we describe novel techniques for querying the change. Using the example of this archived curated database, we discuss the extent to which the accepted practices and terminology of archiving, curation and digital preservation apply to this important class of digital artefacts.1 1 This paper is based on the paper given by the authors at the 5th International Digital Curation Conference, December 2009; received November 2009, published December 2009. -

Understanding the Israeli-Palestinian Conflict
Understanding the Israeli-Palestinian Conflict Global Classroom Workshops made possible by: THE Photo Courtesy of Bill Taylor NORCLIFFE FOUNDATION A Resource Packet for Educators Compiled by Kristin Jensen, Jillian Foote, and Tese Wintz Neighbor And World May 12, 2009 Affairs Council Members HOW TO USE THIS RESOURCE GUIDE Please note: many descriptions were excerpted directly from the websites. Packet published: 5/11/2009; Websites checked: 5/11/2009 Recommended Resources Links that include… Lesson Plans & Charts & Graphs Teacher Resources Audio Video Photos & Slideshows Maps TABLE OF CONTENTS MAPS 1 FACT SHEET 3 TIMELINES OF THE CONFLICT 4 GENERAL RESOURCES ON THE ISRAELI-PALESTINIAN CONFLICT 5 TOPICS OF INTEREST 7 CURRENT ARTICLES/EDITORIALS ON THE ISRAELI-PALESTINIAN CONFLICT 8 (Focus on International Policy and Peace-Making) THE CRISIS IN GAZA 9 RIPPED FROM THE HEADLINES: WEEK OF MAY 4TH 10 RELATED REGIONAL ISSUES 11 PROPOSED SOLUTIONS 13 ONE-STATE SOLUTION 14 TWO-STATE SOLUTION 14 THE OVERLAPPING CONUNDRUM – THE SETTLEMENTS 15 CONFLICT RESOLUTION TEACHER RESOURCES 15 MEDIA LITERACY 17 NEWS SOURCES FROM THE MIDEAST 18 NGOS INVOLVED IN ISRAELI-PALESTINIAN RELATIONS 20 LOCAL ORGANIZATIONS & RESOURCES 22 DOCUMENTARIES & FILMS 24 BOOKS 29 MAPS http://johomaps.com/as/mideast.html & www.cia.gov/library/publications/the-world-factbook/geos/is.html Other excellent sources for maps: From the Jewish Virtual Library - http://www.jewishvirtuallibrary.org/jsource/History/maptoc.html Foundation for Middle East Peace - http://www.fmep.org/maps/ -

The Seven States of the Former Yugoslavia: an Evaluation
The Seven States of the Former Yugoslavia: An Evaluation Thomas P. Melady, Ph.D. Professor and Senior Diplomat in Residence, Institute of World Politics United States Ambassador to the Holy See, 1989-1993 United States Ambassador to Uganda, 1972-1973 United States Ambassador to Burundi, 1969-1972 Senior Advisor to the US Delegation to the United Nations General Assembly President Emeritus of Sacred Heart University Former United States Assistant Secretary for Post Secondary Education J. Cushman Laurent Candidate for Master’s Degree, Institute of World Politics Executive Assistant to the Sr. Diplomat in Residence, Institute of World Politics he area formerly known as Yugoslavia, positioned at the crossroads of East and West, is a T melting pot of ethnicities and religions. As one country, Yugoslavia’s rich multi- culturalism was a source of contention, culminating in a series of bloody conflicts in the early 1990s. The Dayton Accords of 1995 brought peace to the region and created separate nations organized along ethnic and religious lines. Sixteen years after the signing of the Dayton Accords, we examine the geo- political situation in each of the seven independent states of the Source: Encyclopedia Britannica Online, 2011. former Yugoslavia. Bosnia and Herzegovina In the early 1990s, there was considerable ethnic-religious conflict in Bosnia and Herzegovina. The Dayton Peace Accords had the immediate impact of bringing the inter- ethnic strife to an end. Bosnia and Herzegovina was confirmed as a sovereign state. Today, the country is slightly smaller than West Virginia and has a population of over four and a 1 half million. -

Mapping the Global Muslim Population
MAPPING THE GLOBAL MUSLIM POPULATION A Report on the Size and Distribution of the World’s Muslim Population October 2009 About the Pew Forum on Religion & Public Life This report was produced by the Pew Research Center’s Forum on Religion & Public Life. The Pew Forum delivers timely, impartial information on issues at the intersection of religion and public affairs. The Pew Forum is a nonpartisan, nonadvocacy organization and does not take positions on policy debates. Based in Washington, D.C., the Pew Forum is a project of the Pew Research Center, which is funded by The Pew Charitable Trusts. This report is a collaborative effort based on the input and analysis of the following individuals: Pew Forum on Religion & Public Life Communications and Web Publishing Luis Lugo, Director Erin O’Connell, Associate Director, Communications Oliver Read, Web Manager Research Loralei Coyle, Communications Manager Alan Cooperman, Associate Director, Research Robert Mills, Communications Associate Brian J. Grim, Senior Researcher Liga Plaveniece, Program Coordinator Mehtab S. Karim, Visiting Senior Research Fellow Sahar Chaudhry, Research Analyst Pew Research Center Becky Hsu, Project Consultant Andrew Kohut, President Jacqueline E. Wenger, Research Associate Paul Taylor, Executive Vice President Kimberly McKnight, Megan Pavlischek and Scott Keeter, Director of Survey Research Hilary Ramp, Research Interns Michael Piccorossi, Director of Operations Michael Keegan, Graphics Director Editorial Alicia Parlapiano, Infographics Designer Sandra Stencel, Associate Director, Editorial Russell Heimlich, Web Developer Andrea Useem, Contributing Editor Tracy Miller, Editor Sara Tisdale, Assistant Editor Visit http://pewforum.org/docs/?DocID=450 for the interactive, online presentation of Mapping the Global Muslim Population. -
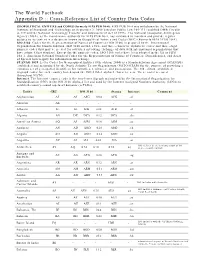
The World Factbook Appendix D :: Cross-Reference List of Country Data Codes
The World Factbook Appendix D :: Cross-Reference List of Country Data Codes GEOPOLITICAL ENTITIES and CODES (formerly FIPS PUB 10-4): FIPS PUB 10-4 was withdrawn by the National Institute of Standards and Technology on September 2, 2008 based on Public Law 104-113 (codified OMB Circular A-119 and the National Technology Transfer and Advancement Act of 1995). The National Geospatial-Intelligence Agency (NGA), as the maintenance authority for FIPS PUB 10-4, has continued to maintain and provide regular updates to its content in a document known as Geopolitical Entities and Codes (GEC) (Formerly FIPS 1PUB 10-4). ISO 3166: Codes for the Representation of Names of Countries (ISO 3166) is prepared by the International Organization for Standardization. ISO 3166 includes two- and three-character alphabetic codes and three-digit numeric codes that may be needed for activities involving exchange of data with international organizations that have adopted that standard. Except for the numeric codes, ISO 3166 codes have been adopted in the US as FIPS 104-1: American National Standard Codes for the Representation of Names of Countries, Dependencies, and Areas of Special Sovereignty for Information Interchange. STANAG 1059: Letter Codes for Geographical Entities (8th edition, 2004) is a Standardization Agreement (STANAG) established and maintained by the North Atlantic Treaty Organization (NATO/OTAN) for the purpose of providing a common set of geo-spatial identifiers for countries, territories, and possessions. The 8th edition established trigraph codes for each country based upon the ISO 3166-1 alpha-3 character sets. These codes are used throughout NATO. Internet: The Internet country code is the two-letter digraph maintained by the International Organization for Standardization (ISO) in the ISO 3166 Alpha-2 list and used by the Internet Assigned Numbers Authority (IANA) to establish country-coded top-level domains (ccTLDs). -

The World Factbook
The World Factbook Europe :: Slovakia Introduction :: Slovakia Background: Slovakia's roots can be traced to the 9th century state of Great Moravia. Subsequently, the Slovaks became part of the Hungarian Kingdom, where they remained for the next 1,000 years. Following the formation of the dual Austro-Hungarian monarchy in 1867, language and education policies favoring the use of Hungarian (Magyarization) resulted in a strengthening of Slovak nationalism and a cultivation of cultural ties with the closely related Czechs, who were under Austrian rule. After the dissolution of the Austro-Hungarian Empire at the close of World War I, the Slovaks joined the Czechs to form Czechoslovakia. During the interwar period, Slovak nationalist leaders pushed for autonomy within Czechoslovakia, and in 1939 Slovakia became an independent state allied with Nazi Germany. Following World War II, Czechoslovakia was reconstituted and came under communist rule within Soviet-dominated Eastern Europe. In 1968, an invasion by Warsaw Pact troops ended the efforts of the country's leaders to liberalize communist rule and create "socialism with a human face," ushering in a period of repression known as "normalization." The peaceful "Velvet Revolution" swept the Communist Party from power at the end of 1989 and inaugurated a return to democratic rule and a market economy. On 1 January 1993, the country underwent a nonviolent "velvet divorce" into its two national components, Slovakia and the Czech Republic. Slovakia joined both NATO and the EU in the spring of 2004 -

1 of 8 7/26/16, 7:42 PM
https://www.cia.gov/library/publications/the-world-factbook/fiel... FIELD LISTING :: SUFFRAGE This entry gives the age at enfranchisement and whether the right to vote is universal or restricted. Country Comparison to the World COUNTRY SUFFRAGE Afghanistan 18 years of age; universal Albania 18 years of age; universal Algeria 18 years of age; universal American Samoa 18 years of age; universal Andorra 18 years of age; universal Angola 18 years of age; universal Anguilla 18 years of age; universal Antigua and Barbuda 18 years of age; universal Argentina 18-70 years of age; universal and compulsory; 16-17 years of age - optional for national elections Armenia 18 years of age; universal Aruba 18 years of age; universal Australia 18 years of age; universal and compulsory Austria 16 years of age; universal Azerbaijan 18 years of age; universal Bahamas, The 18 years of age; universal Bahrain 20 years of age; universal; note - Bahraini Cabinet in May 2011 endorsed a draft law lowering eligibility to 18 years Bangladesh 18 years of age; universal Barbados 18 years of age; universal Belarus 18 years of age; universal Belgium 18 years of age; universal and compulsory Belize 18 years of age; universal Benin 18 years of age; universal Bermuda 18 years of age; universal Bhutan 18 years of age; universal Bolivia 18 years of age, universal and compulsory Bosnia and Herzegovina 18 years of age, 16 if employed; universal Botswana 18 years of age; universal 1 of 8 7/26/16, 7:42 PM https://www.cia.gov/library/publications/the-world-factbook/fiel.. -

The Landpower Essay Series
THE LANDPOWER ESSAY SERIES LPE 20-1 / JANUARY 2020 • PUBLISHED BY THE ASSOCIATION OF THE UNITED STATES ARMY The Strategy of a Great Power Competitor by Captain Johannes Geist, U.S. Army Introduction The following discourse offers an assessment of China’s grand strategy and what the grand strategy of the United States should be toward China, first by defining the term and introducing the concept of the in- struments of national power. With these general ideas established, it is possible to understand the particular grand strategies of China and the United States, allowing the defense community to draw conclusions regard- ing future action that the United States should undertake in its own grand strategy to counter that of China. The Concept of Grand Strategy The first line of thought central to developing an understanding of Chinese interests is establishing a base- line characterization of the concept of grand strategy. To start, the exact confines of grand strategy are con- stantly debated; no one definition is accepted by all. However, Hal Brands, an American historian and author of The Promise and Pitfalls of Grand Strategy, offers that “British military historian Sir Basil Liddell Hart popularized the term during the mid-20th century, and most subsequent definitions of the concept have been based, in one way or another, on his insight that grand strategy involves synchronizing means and ends at the highest level of national policy.” Brands says “grand strategy is defined as the theory, or logic, that binds a country’s highest interests to its daily interactions with the world.