(Solanum Lycopersicum Mill.) by SABIR HUSSAIN
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
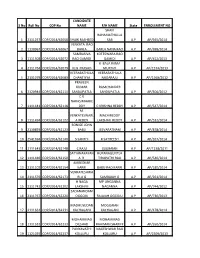
S No Roll No COP No CANDIDATE NAME F/H NAME State
CANDIDATE S No Roll No COP No NAME F/H NAME State ENROLLMENT NO SHAIK RAHAMATHULLA 1 2111257 COP/2014/62058 SHAIK RASHEED SAB A.P AP/945/2014 VENKATA RAO 2 1130967 COP/2014/62067 BARLA BARLA NANA RAO A.P AP/698/2014 SAMBASIVA KOTESWARA RAO 3 1111308 COP/2014/62072 RAO GAMIDI GAMIDI A.P AP/452/2013 K BALA RAMA 4 2111764 COP/2014/62079 KLN PRASAD MURTHY A.P AP/1574/2013 VEERABATHULA VEERABATHULA 5 2131079 COP/2014/62083 CHANTIYYA NAGARAJU A.P AP/1568/2012 PRAVEEN KUMAR RAMCHANDER 6 2120944 COP/2014/62111 SANDUPATLA SANDUPATLA A.P AP/306/2012 C V NARASIMHARE 7 1111441 COP/2014/62118 DDY C KRISHNA REDDY A.P AP/547/2014 M. VENKATESWARL MACHIREDDY 8 1111494 COP/2014/62122 A REDDY LAKSHMI REDDY A.P AP/532/2014 BONIGE JOHN 9 2130893 COP/2014/62123 BABU JEEVARATNAM A.P AP/878/2014 10 2541694 COP/2014/62140 S SANTHI R SATHEESH A.P AP/267/2014 11 2111643 COP/2014/62148 C RAJU SUGRAIAH A.P AP/1238/2011 SATYANARAYAN RUPANAGUNTLA 12 1111480 COP/2014/62150 A R. TIRUPATHI RAO A.P AP/540/2014 AMBEDKAR 13 2131102 COP/2014/62154 KARRI BABU RAO KARRI A.P AP/180/2014 VENKATESHWA 14 2111570 COP/2014/62173 RLU G SAMBAIAH G A.P AP/261/2014 H NAGA MP LINGANNA 15 2111742 COP/2014/62202 LAKSHMI NAGANNA A.P AP/744/2012 SADANANDAM 16 2111767 COP/2014/62220 OGGOJU RAJAIAH OGGOJU A.P AP/736/2013 MADHUSUDAN MOGILAIAH 17 2111661 COP/2014/62231 KACHAGANI KACHAGANI A.P AP/478/2014 MOHAMMAD MOHAMMAD 18 1111532 COP/2014/62233 DILSHAD RAHIMAN SHARIFF A.P AP/550/2014 PUNYAVATHI NAGESHWAR RAO 19 1121035 COP/2014/62237 KOLLURU KOLLURU A.P AP/2309/2013 G SATHAKOTI GEESALA 20 2131021 COP/2014/62257 SRINIVAS NAGABHUSHANAM A.P AP/1734/2011 GANTLA GANTLA SADHU 21 1131067 COP/2014/62258 SANYASI RAO RAO A.P AP/1802/2013 KOLICHALAM NAVEEN KOLICHALAM 22 1111688 COP/2014/62265 KUMAR BRAHMAIAH A.P AP/1908/2010 SRINIVASA RAO SANKARA RAO 23 2131012 COP/2014/62269 KOKKILIGADDA KOKKILIGADDA A.P AP/793/2013 24 2120971 COP/2014/62275 MADHU PILLI MAISAIAH PILLI A.P AP/108/2012 SWARUPARANI 25 2131014 COP/2014/62295 GANJI GANJIABRAHAM A.P AP/137/2014 26 2111507 COP/2014/62298 M RAVI KUMAR M LAXMAIAH A.P AP/177/2012 K. -

Pachmarhi Cantt Area Ward No. 01(YEAR 2020-2021) SEE RULE 11
Pachmarhi Cantt Area Ward No. 01(YEAR 2020-2021) SEE RULE 11 S.No. NAME FATHER/HUSBAND YEAR ADDRESS NAME 1. SITA CHARAN S/O VANSHI PRASAD 79 93/185 JAWAHAR GANJ 2. LATA W/O SITA CHARAN 69 93/185 JAWAHAR GANJ 3. MANISH S/O SITA CHARAN 48 93/185 JAWAHAR GANJ 4. SHILU W/O MANISH 43 93/185 JAWAHAR GANJ 5. SUKHLAL S/O DHANRAJ 93/185 JAWAHAR PRANDRE 18 GANJ 6. HEMRAJ S/O JAGDISH PATWA 93/185 JAWAHAR 19` GANJ 7. SUSHIL PALIWAL S/O PYARE LAL 64 93/185 JAWAHAR GANJ 8. SAVITRI W/O SUSHIL 53 93/185 JAWAHAR GANJ 9. SATISH S/O SUSHIL 30 93/185 JAWAHAR GANJ 10. DINESH MALANI S/O TULSI RAM 65 93/185 JAWAHAR GANJ 11. BINA MALANI W/O DINESH 61 93/185 JAWAHAR GANJ 12. GYANESH S/O GIRDHARILAL 58 93/184 JAWAHAR GANJ 13. MOTI LAL S/O RAMPRASAD 81 93/182 JAWAHAR GANJ 14. SHOBHA RANI W/O MOTI LAL 73 93/182 JAWAHAR GANJ 15. DEEPAK S/O MOTILAL 52 93/182 JAWAHAR GANJ 16. SANGEETA W/O DEEPAK 49 93/182 JAWAHAR GANJ 17. SANDEEP S/O MOTI LAL 48 93/182 JAWAHAR GANJ 18. KANCHAN W/O SANDEEP 47 93/182 JAWAHAR GANJ Page 1/33 Pachmarhi Cantt Area Ward No. 01(YEAR 2020-2021) SEE RULE 11 S.No. NAME FATHER/HUSBAND YEAR ADDRESS NAME 19. NUPUR D/O DEEPAK 25 93/182 JAWAHAR GANJ 19. HARSHITA D/O DEEPAK 19 93/182 JAWAHAR 20. K.B. -

General Awareness–Current Affairs Month of March-2019
GENERAL AWARENESS–CURRENT AFFAIRS MONTH OF MARCH-2019 List of Important Days March 1 - Zero Discrimination Day (Theme – “Act to change laws that Discriminate”) March 4 - National Safety Day (Themes – “Cultivate and Sustain A Safety Culture for Building Nation”) Mar 4-10 - National Safety Week March 7 - Janaushadhi Diwas March 8 - International Women’s Day (Theme – “Think Equal, Build Smart, Innovate for Change”). March 12 - World Day against Cyber Censorship March 12 - 30th anniversary of the World Wide Web (WWW) March 14 - (2nd Thursday of March) World Kidney Day (Theme - “Kidney Health for Everyone Everywhere”) March 14 - Pi Day (Pi's value (3.14)) March 15 - World Consumer Rights Day (In India this day is celebrated as Viswa Upabhokta Adhikar Diwas). (Theme – “Trusted Smart Products”) March 20 - International Day of Happiness. (Theme – “Happier Together”) March 20 - World Day of Theatre for Children and Young People March 20 - World Sparrow Day. (Theme – “I LOVE Sparrows”) March 21 - International Day of Forests. (Theme “Forests and Education”) March 21 - World Poetry Day March 21 - World Down Syndrome Day March 21 - International Day for the Elimination of Racial Discrimination (Theme – “Mitigating and countering rising nationalist populism and extreme supremacist ideologies”) March 21 - World Puppetry Day March 22 - World Water Day (Theme – “Leaving no one behind”) March 23 - World Meteorological Day (Theme – “The Sun, the Earth and the Weather”) March 23 - 88th Shaheed Diwas (Martyr’s Day) March 24 - World Tuberculosis (TB) Day (Theme – “It’s time”) March 25 - International Day of Remembrance of the Victims of Slavery and Transatlantic Slave Trade. (Theme – “Remember Slavery: The Power of the Arts for Justice”) March 26 - Independence Day of Bangladesh March 27 - World Theatre Day (WTD) March 30 - Rajasthan Diwas Reserve Bank of India • The Reserve Bank of India (RBI) has fined Yes Bank ₹1 crore for not complying with its directions about SWIFT, a financial messaging software. -

BOOTH-1 (Sr.No-01 to 1045) ALPHABET-A to M BOOTH-1 VOTER LIST of WORKSHOP-CRWS/BPL AS on 31/01/2020 Sr
VOTER’S LIST CRWS BHOPAL BOOTH-1 (Sr.No-01 to 1045) ALPHABET-A to M BOOTH-1 VOTER LIST OF WORKSHOP-CRWS/BPL AS ON 31/01/2020 Sr. Emp No. Emp Name Father Name Designation Department Work Signature No. Place 1 45504158830 A.K. GAUTAM KISHORI LAL TECH.CARPEN MECHANICAL BODY GAUTAM T ER-I 2 45504159184 A.K.AWASTHI SITARAM TECH.(ELEC)-I ELECTRICAL ETL AWASTHI 3 45317563290 A.K.JHA SHRI I.D.JHA Sr.CASHIER ACCOUNTS ACC 4 45505825441 AAJAD MEHRA S L MEHRA TECH(PAINTER MECHANICAL BODY )- II 5 45508383789 ABDUL ANWAR ABDUL MAJEED Sr.TECH(FITTE MECHANICAL BOGIE R) 6 45506146703 ABDUL HAMEED ABDUL GAFUR TECH.CARPEN MECHANICAL FUR T ER-I 7 45500963446 ABDUL ISRAR ABDUL BASHIR TECH.CARPEN MECHANICAL CARP T ER-I 8 45505823559 ABDUL MATEEN KHAN ABDUL BAHID Sr.TECH. MECHANICAL FUR KHAN CARPENTER 9 45508216642 ABDUL MATIN ABDUL LATIF TECH.(TL)-I ELECTRICAL ETL 10 45507471191 ABDUL RASHEED KHAN A R KHAN Sr.TECH.(TL) ELECTRICAL ETL 11 45507465075 ABDUL RAZZAK GULAB KHAN WELDER-I MECHANICAL PCO 12 45505823523 ABDUL REHMAN KHAN ABDUL WAHEED TECH.CARPEN MECHANICAL BOGIE KHAN T ER-I 13 45505824497 ABDUL SATTAR SHEKH NATHU Sr.TECH(FITTE MECHANICAL BODY R) 14 45508204949 ABHIJEET DESHMUKH SHRI SUBHASH AA ACCOUNTS ACC DESHMUKH 15 45529800157 ABHIMANYU RAIKWAR MUNNA LAL JR CLERK PERSONNEL PER RAIKWAR 16 45505827231 ABHIMANYU SINGH DHARAM PAL ASST(WS) MECHANICAL FUR PATEL 17 45516641114 ABHINAY KUMAR SONI JAMUNA SONI ASST(WS) MECHANICAL BODY 18 45529800005 ABHISHEK CHAUHAN LATE MAHESH Sr. CLERK PERSONNEL PER KUMAR 19 45529800024 ABHISHEK KUMAR JAI BHAGWAN ASST(WS) MECHANICAL BODY SINGH SINGH 20 45529800013 ABHISHEK PADERIYA SHRI MISHRI LAL ASST STORES GSD LODHI DEPOT(STR) 21 45529800136 ABHISHEK SINGH BANSHPATI SSE MECH MECHANICAL MSUP PATEL 22 45505825283 ABHISHEK SONI A P SONI WELDER-III MECHANICAL BODY 23 45505825386 ABHISHEK TIWARI R N TIWARI FITTER-I MECHANICAL N 24 45529800147 ABID KHAN QASIM KHAN TECH.(OP)-II MECHANICAL TRN 25 45505824503 ADHAR SINGH PUNDLIK Sr.TECH(WELD MECHANICAL BODY ) 26 45505823791 AFROZ KHAN RAMZAN KHAN Sr.TECH. -

Nominations for Padma Awards 2011
c Nominations fof'P AWARDs 2011 ADMA ~ . - - , ' ",::i Sl. Name';' Field State No ShriIshwarappa,GurapJla Angadi Art Karnataka " Art-'Cinema-Costume Smt. Bhanu Rajopadhye Atharya Maharashtra 2. Designing " Art - Hindustani 3. Dr; (Smt.).Prabha Atre Maharashtra , " Classical Vocal Music 4. Shri Bhikari.Charan Bal Art - Vocal Music 0, nssa·' 5. Shri SamikBandyopadhyay Art - Theatre West Bengal " 6: Ms. Uttara Baokar ',' Art - Theatre , Maharashtra , 7. Smt. UshaBarle Art Chhattisgarh 8. Smt. Dipali Barthakur Art " Assam Shri Jahnu Barua Art - Cinema Assam 9. , ' , 10. Shri Neel PawanBaruah Art Assam Art- Cinema Ii. Ms. Mubarak Begum Rajasthan i", Playback Singing , , , 12. ShriBenoy Krishen Behl Art- Photography Delhi " ,'C 13. Ms. Ritu Beri , Art FashionDesigner Delhi 14. Shri.Madhur Bhandarkar Art - Cinema Maharashtra Art - Classical Dancer IS. Smt. Mangala Bhatt Andhra Pradesh Kathak Art - Classical Dancer 16. ShriRaghav Raj Bhatt Andhra Pradesh Kathak : Art - Indian Folk I 17., Smt. Basanti Bisht Uttarakhand Music Art - Painting and 18. Shri Sobha Brahma Assam Sculpture , Art - Instrumental 19. ShriV.S..K. Chakrapani Delhi, , Music- Violin , PanditDevabrata Chaudhuri alias Debu ' Art - Instrumental 20. , Delhi Chaudhri ,Music - Sitar 21. Ms. Priyanka Chopra Art _Cinema' Maharashtra 22. Ms. Neelam Mansingh Chowdhry Art_ Theatre Chandigarh , ' ,I 23. Shri Jogen Chowdhury Art- Painting \VesfBengal 24.' Smt. Prafulla Dahanukar Art ~ Painting Maharashtra ' . 25. Ms. Yashodhara Dalmia Art - Art History Delhi Art - ChhauDance 26. Shri Makar Dhwaj Darogha Jharkhand Seraikella style 27. Shri Jatin Das Art - Painting Delhi, 28. Shri ManoharDas " Art Chhattisgarh ' 29. , ShriRamesh Deo Art -'Cinema ,Maharashtra Art 'C Hindustani 30. Dr. Ashwini Raja Bhide Deshpande Maharashtra " classical vocalist " , 31. ShriDeva Art - Music Tamil Nadu Art- Manipuri Dance 32. -

THE RECORD NEWS ======The Journal of the ‘Society of Indian Record Collectors’, Mumbai ------ISSN 0971-7942 Volume – Annual: TRN 2008 ------S.I.R.C
THE RECORD NEWS ============================================================= The journal of the ‘Society of Indian Record Collectors’, Mumbai ------------------------------------------------------------------------ ISSN 0971-7942 Volume – Annual: TRN 2008 ------------------------------------------------------------------------ S.I.R.C. Branches: Mumbai, Pune, Solapur, Nanded, Tuljapur, Baroda, Amravati ============================================================= Feature Article: Gramophone Celebrities-III 1 ‘The Record News’ - Annual magazine of ‘Society of Indian Record Collectors’ [SIRC] {Established: 1990} -------------------------------------------------------------------------------------------- President Narayan Mulani Hon. Secretary Suresh Chandvankar Hon. Treasurer Krishnaraj Merchant ==================================================== Patron Member: Mr. Michael S. Kinnear, Australia -------------------------------------------------------------------------------------------- Honorary Members V. A. K. Ranga Rao, Chennai Harmandir Singh Hamraz, Kanpur -------------------------------------------------------------------------------------------- Membership Fee: [Inclusive of the journal subscription] Annual Membership Rs. 1000 Overseas US $ 100 Life Membership Rs. 10000 Overseas US $ 1000 Annual term: July to June Members joining anytime during the year [July-June] pay the full membership fee and get a copy of ‘The Record News’ published in that year. Life members are entitled to receive all the back issues in five bound volumes -

Islamabad High Court
_ _ ISLAMABAD HIGH COURT DAILY REGULAR CAUSE LIST FOR MONDAY, 27 SEPTEMBER, 2021 THE HONORABLE CHIEF JUSTICE & Court No: 1 BEFORE:- MR. JUSTICE AAMER FAROOQ NOTE: Old cases will not be adjourned except with prior adjustments and with the consent of opposite counsel. OLD CASES 1. Cust. Ref. 2/2015 (46152) Director of Intelligence and Investigation V/s SS Corporation, etc A CM 1/2015 Mujeeb-ur-Rehman Warraich CM 2/2015 MOTION CASES 1. Crl. Appeal 77/2021 Acquittal (131152) The State V/s Muhammad Israr etc A Other Advocate General NOTICE CASES 1. Part Heard (130761) Mosharraf Ali Zaidi & others V/s The President of Pakistan & others FC W.P. 1925/2021 Misc. Other FAISAL SIDDIQI Additional Attorney General, CM 2/2021 Assistant Attorney General, Attorney General for Pakistan, Deputy Attorney General, Mansoor Tariq, Ms.Kulsum Khaliq, Shahid Hamid, Sikandar Naeem Qazi W.P. 127/2021 (125274) Dr. Asfandiyar, etc V/s FOP, etc FC CM 1/2021 (Soban Ali, Adv.) In Person, Mudassar Khalid Moazzam Ali Shah, Qausain Abbasi, Arshad Abbas Faisal Mufti, Assistant Attorney General 2. Crl. Misc. 949/2021 Bail After (134907) Zahir Ullah V/s State, etc A Arrest Muhammad Shaheen By I.T. Department, Islamabad High Court Report Auto Generated By: C F M S Print Date & Time:23-SEP-2021 05:23 PM Page 1 of 87 _ _ DAILY REGULAR CAUSE LIST FOR MONDAY, 27 SEPTEMBER, 2021 THE HONORABLE CHIEF JUSTICE & Court No: 1 BEFORE:- MR. JUSTICE AAMER FAROOQ NOTICE CASES 3. Crl. Appeal 208/2020 Against (124285) Bait Ullah V/s The State etc FC Convct. -

Unpaid Warrants 31.03.2015
CAREER POINT LIMITED Registered & Corporate Office: CP Tower 1, Road No. 1, IPIA, Kota, Rajasthan 324005 Corporate Identification Number: L72200RJ2000PLC016272, Phone: 0744-3040000, 0744-3040050, www.cpil.in, [email protected] Interim Dividend -DD Cancelled and Credited-As on 31st March 2015 AMOUNT NAME 20.00 VISHASHER LAL SETIA 20.00 GURCHARAN JIT SINGH 20.00 AMIT VERMA 20.00 HARI RAM GUPTA 10.00 BASANT KUMAR HAZRATI 20.00 CHANDRA PRAKASH KALANTRI 15.00 MANISH KHATRI 10.00 HETALBEN ARVINDBHAI PATEL 1.00 MAYUR JAMNADAS VORA 5.00 ALKA PRABHAT GUPTA 20.00 DEVICA NANIK CHANDIRAMANI 5.00 KRISHNAKUMAR RAJGOPAL TOSHNIWAL 20.00 Sujit Ramkrishna Jajoo 20.00 NAMRATA CHANDRASHEKHAR MESHRAM 50.00 PANDIT SHANKARRAO PACHPANDE 3.00 ANIL MADANLAL PANPALIYA 2,427.00 KARVY STOCK BROKING LTD 50.00 RAVI AGGARWAL 5.00 INDER KUMAR VASUDEVA 100.00 PUNEET GUPTA 70.00 S C GULATI 20.00 KRISHNA VERMA 20.00 MANOJ KUMAR AGARWAL 6.00 HAWA SINGH 2.00 KAILASH 20.00 SUNIL KUMAR 20.00 RAM NIWAS YADAV 100.00 GEETA 40.00 SANDEEP MITTAL 329.00 PRATIMA 8.00 MANJEET SINGH 100.00 NEELAM BHATIA 200.00 JOGINDER SINGH 100.00 DAVINDER MALIK 6.00 RAJAN BAJAJ 78.00 PAWAN KUKAR 50.00 MADHU BALHOTRA 7.00 SUBASH CHANDER SHARMA 93.00 SANJAY SHARMA 7.00 NISAR AHMED KHANDAY 50.00 RAVINDRA KUMAR KASANA 5.00 SEEMA GOYAL 220.00 SAIYED MOHAMMED ALI BADRUDDIN 20.00 RITU RANI 20.00 SANJAY BHASIN 200.00 ARUN PRAKASH AGRAWAL 200.00 GOVIND MAHESHWARI 20.00 SUNIL KUMAR VARMA 10.00 PUSPA KARWARIYA 15.00 SAMIR BHASIN 10.00 GARIMA ARORA 50.00 AKBAR ALI KHAN 50.00 KAVITA SINGH 44.00 VINOD KUMAR MAURYA -
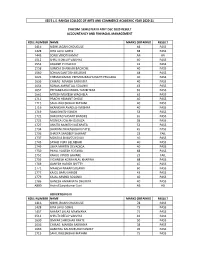
Roll Number Name Marks Obtained Result 1416 Nidhi
SES'S L.S. RAHEJA COLLEGE OF ARTS AND COMMERCE ACADEMIC YEAR 2020-21 SYBCOM SEMESTER III ATKT DEC 2020 RESULT ACCOUNTANCY AND FINNACIAL MANAGEMENT ROLL NUMBER NAME MARKS OBTAINED RESULT 1416 NIDHI JAGAN CHOUGULE 68 PASS 1428 RITA LAVJI GOHIL 68 PASS 1443 SONU VINOD KAMAT AB AB 1512 SHRUTI DEELIP VAISHYA 40 PASS 1552 ADARSH P CHACKO 43 PASS 1558 AJINKYA SHANKAR BACHCHE 40 PASS 1560 SOHAN SANTOSH BELOSHE 48 PASS 1623 KHEMAKUMARI PREMRAJRAGHUNATH PHULARA 40 PASS 1636 CHIRAG MANISH SARVAIYA 40 PASS 1654 RONAK AMRUTLAL SOLANKI 43 PASS 1657 PRIYANKA RAVINDRA TAMBITKAR 65 PASS 1661 MITESH MUKESH WAGHELA 63 PASS 1711 PRACHI HEMANT DHOLE 40 PASS 1715 SAHIL RAJESHBHAI MAYANI 40 PASS 1716 AKANKSHA AVADESH MISHRA 40 PASS 1719 RANI DINESH SINGH 53 PASS 1722 DHRUPAD VASANT BANDRE 65 PASS 1723 MONICA JOSEPH DSOUZA 58 PASS 1727 ANKITA MUKESH KATAPATA 53 PASS 1734 DHWANI PRAKASHBHAI PATEL 45 PASS 1736 SHREYA SANDEEP SAWANT 23 FAIL 1737 MOKSHA BHAVESH SHAH 40 PASS 1743 SAYALI VIJAY BELNEKAR 40 PASS 1749 LIBYA MARTIN DEVADIGA 40 PASS 1750 PAYAL YOGESH FOTARIA 68 PASS 1751 RAHUL VINOD GAVARE 23 FAIL 1756 VIGHNESH KONAYALAL KHARWA 68 PASS 1769 GANESH HARISH SHETTY 45 PASS 1771 MAHESH NARAN SOLANKI 60 PASS 1777 KAJOL BAPU SHINDE 43 PASS 1779 KAJAL ARVIND SOLANKI 40 PASS 1786 GANESH AMARNATH DHURIYA 40 PASS A880 Arvind Suryakumar Soni AB AB ADVERTISING III ROLL NUMBER NAME MARKS OBTAINED RESULT 1416 NIDHI JAGAN CHOUGULE 78 PASS 1428 RITA LAVJI GOHIL 73 PASS 1457 BHARAT LALAJI MAKWANA 75 PASS 1512 SHRUTI DEELIP VAISHYA 43 PASS 1630 OMKAR SHRIDHAR RAKTE 50 PASS -

Placement Details of January 2012 to March 2013
List of Placed Candidates January 2012 to March 2013 January Selection - 2012 Sl No Name Trade Remark 1 Nand Lal Pal Land Surveyor Selected by ARSS.Infrastructure Project Ltd.8000/ P. Month + Accomodation 2 Roshan Khan 3 Jalal Uddin Hazam GWS Selected by ARSS.Infrastructure Project Ltd.8000/ P. Month + Accomodation 4 Sahid Hussain Laskar Storekeeper 5 Gaurav Kumar GWS 6 Jai Shankar Maurya GWS 7 Rajesh Kumar Verma GWS 8 Jagannath GWS 9 Hari Shankar Pal GWS 10 Prabhat Gupta GWS 11 Indroj Kumar S GWS 12 Rakesh Kumar Verma GWS 13 Vinay Kumar Pal GWS 14 Mithun Rabidas GWS 15 Shiv Raj Singh GWS 16 Gaurav Kumar Singh GWS Selected by ERA Group, 6500/ P. Month+ Food + Accomodation 17 Rabi Kalindi GWS 18 Amir Suhel Laskar GWS 19 Priotush Dey GWS 20 Pawan Kumar Land Surveyor 21 Mahabeer Yadav Land Surveyor 22 Idrish Khan Land Surveyor 23 Shah Faisal Land Surveyor 24 Abbas Khan Land Surveyor 25 Nitesh Kumar Land Surveyor 26 Sadik Hussain Land Surveyor 27 Joginder Singh Land Surveyor 28 Umesh Kumar GWS Selected by Era Group, 6000/P. Month + Accomodation 29 Pinak Roy BMLT 30 Sahid Ahmad Choudhury BMLT Selected by ERA Group, 6500/P. Month+ Food + Accomodation P. Month 31 Sandip Dutta Mazumdar Electrician 32 Prem Chandra Maurya GWS 33 Ramesh Chandra GWS 34 Deepak Kumar GWS 35 Jitendra Nigam GWS Selected By Shobha Developers Ltd. 7150/ P. Month+ Accomodation. 36 Nagendra Singh Gautam GWS 37 Avinash Chandra pandey GWS 38 Kamal Prasad Kairi GWS February Selection - 2012 1 Khuli Mohd GWS 2 Nadeem Anwar GWS 3 Mohd Ajij GWS 4 Sabir Khan GWS 5 Saddam Hussain GWS 6 Imran Khan GWS Selected by BLG Contuation Service (P) Ltd. -

Primo.Qxd (Page 1)
SATURDAY, NOVEMBER 16, 2013 (PAGE 12) DAILY EXCELSIOR, JAMMU (From page-11) 33 317024573 MUZAMIL WAHEED AB WAHEED WANI 27 November 2013 166 318027247 NAZAR MOHAMMED SHER MOHAMMED 4 December 2013 134 321029246 PERVEZ AHMAD KHAN AB RASHID KHAN 27 November 2013 34 317024574 GOURAV TALWAR SHAM LAL TALWAR 27 November 2013 167 318027248 DHEERAJ THAKUR SURAJ SINGH 4 December 2013 135 321029253 LALIT KUMAR MADAN LAL 27 November 2013 6 315021291 JAHANGEER RASOOL GH RASOOL BHAT 18 November 2013 35 317024585 PAYAR SINGH MATHER SINGH 27 November 2013 168 318027254 AMAN SHARMA VIJAY KUMAR 4 December 2013 136 321029254 MANZOOR AHMAD DAR MOHD SUBHAN DAR 27 November 2013 7 315021292 ARSHID AHMAD NAJAR MOHAMMAD SHABAN NAJAR 18 November 2013 36 317024592 MOHD IKHLAQ BASHIR AHMED MALIK 27 November 2013 169 318027256 ASHWANI KUMAR ATRI RAM LAL 4 December 2013 137 321029260 HILAL AHMAD BABA GH MOHD BABA 27 November 2013 8 315021295 REYAZ AHMAD BABA BASHIR AHAMD BABA 18 November 2013 37 317024605 URPHY AHMED MOHD YOUSAF BEG 27 November 2013 170 318027257 ANKUSH SHARMA NARINDER MOHAN SHARMA 4 December 2013 138 321029267 ASHOK DOGRA S.H GAIN CHNAD 27 November 2013 9 315021296 SHAHBAZ MAQBOOL MOHD MAQBOOL BHAT 18 November 2013 38 317024629 OMESH SINGH SIBRAN SINGH 27 November 2013 171 318027258 SAFUL UD DIN NAZIR AHMED 4 December 2013 139 321029272 ABID AHMAD WANI GHULAM MOHAMMAD WANI 27 November 2013 10 315021297 SAJD AHMAD BHAT MOHD YOUSUF BHAT 18 November 2013 39 317024650 MUBARAK HUSSAIN DAING GHULAM HUSSAIN DAING 27 November 2013 172 318027261 JERNAIL SINGH -

Sr.No. Application No. Student Name / Father's Name 1 1523075
Sr.No. Application No. Student Name / Father's Name 1 1523075 RAJDEEP KAUR / JASVEER SINGH 2 1267670 HAFSA / MOHAMMAD RAMZAN 3 1261966 SHAHIDA PARVEEN / BHOLA KHAN 4 1269164 AMANPREET SINGH / HARBANS SINGH 5 1261054 ANMOLPREET SINGH / SATPAL SINGH 6 1294115 ANMOLPREET KAUR / INDERJIT SINGH 7 1269033 ARSHDEEP KAUR / SARABJIT SINGH 8 1261040 SUMIT KUMAR DWIVEDI / CHHOTE LAL 9 1268556 AMRINDER SINGH / FAKKAR SINGH 10 1265743 KESHAV JAIN / SUBHASH JAIN 11 1264284 GURPREET KAUR / BHIM SINGH 12 1266045 NAMAN JAIN / ASHWANI KUMAR 13 1267378 JASMEEN KAUR / SARBJIT SINGH 14 1271813 RAMANPREET KAUR / MANJEET SINGH 15 1266088 DINESH BADHAN / RAM SARUP BADHAN 16 1271965 KARANVIR SINGH / SARABJIT SINGH 17 1260942 KOMALPREET SINGH / HARJIT SINGH 18 1300433 HARAM / LIAQAT ALI 19 1260889 IQRA / MOHAMMAD NASAR 20 1261046 HIMANSHU ARORA / AMIT ARORA 21 1259388 SUMANDEEP KAUR / BHAGWANT SINGH 22 1262315 UPASANA KAUR / GURPARTAP SINGH MOHI 23 1294306 SHABEENA / ABDUL LATIF 24 1275258 HARMANJOT KAUR / RAGHVIR SINGH 25 1261036 ROHIT HANS / JEEVAN KUMAR 26 1267375 DILPREET KAUR / MAJOR SINGH 27 1261043 MANNAN MEHMOOD / MEHMOOD AHMED 28 1260559 MOHD QASIM / MOHD ASHRAF 29 1266721 SATNAM SINGH / BALWINDER SINGH 30 1260697 MOHD SHAHID / MOHD YOUNAS 31 1280336 AKASHDEEP SINGH / YADWINDER SINGH 32 1263281 SARPREET SINGH / KULWINDER SINGH 33 1267505 SAHIL / GAMA KHAN 34 1259511 PARNEET KAUR / JAGTAR SINGH 35 1300414 PRABHJOT SINGH / BAHADAR SINGH 36 1281383 DAVINDER KAUR / LAKHVIR SINGH 37 1294290 SANDEEP KAUR / GURMAIL SINGH 38 1294292 HARDEEP KAUR / PARMJIT SINGH