Wolbachia, an Intracellular Symbiont, Manipulates Bacterial Diversity in Its Insect Host
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The 2014 Golden Gate National Parks Bioblitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event
National Park Service U.S. Department of the Interior Natural Resource Stewardship and Science The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 ON THIS PAGE Photograph of BioBlitz participants conducting data entry into iNaturalist. Photograph courtesy of the National Park Service. ON THE COVER Photograph of BioBlitz participants collecting aquatic species data in the Presidio of San Francisco. Photograph courtesy of National Park Service. The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 Elizabeth Edson1, Michelle O’Herron1, Alison Forrestel2, Daniel George3 1Golden Gate Parks Conservancy Building 201 Fort Mason San Francisco, CA 94129 2National Park Service. Golden Gate National Recreation Area Fort Cronkhite, Bldg. 1061 Sausalito, CA 94965 3National Park Service. San Francisco Bay Area Network Inventory & Monitoring Program Manager Fort Cronkhite, Bldg. 1063 Sausalito, CA 94965 March 2016 U.S. Department of the Interior National Park Service Natural Resource Stewardship and Science Fort Collins, Colorado The National Park Service, Natural Resource Stewardship and Science office in Fort Collins, Colorado, publishes a range of reports that address natural resource topics. These reports are of interest and applicability to a broad audience in the National Park Service and others in natural resource management, including scientists, conservation and environmental constituencies, and the public. The Natural Resource Report Series is used to disseminate comprehensive information and analysis about natural resources and related topics concerning lands managed by the National Park Service. -

1Gyd Lichtarge Lab 2006
Pages 1–7 1gyd Evolutionary trace report by report maker January 18, 2010 4.3.3 DSSP 6 4.3.4 HSSP 6 4.3.5 LaTex 6 4.3.6 Muscle 6 4.3.7 Pymol 6 4.4 Note about ET Viewer 6 4.5 Citing this work 6 4.6 About report maker 7 4.7 Attachments 7 1 INTRODUCTION From the original Protein Data Bank entry (PDB id 1gyd): Title: Structure of cellvibrio cellulosa alpha-l-arabinanase Compound: Mol id: 1; molecule: arabinan endo-1,5-alpha-l- arabinosidase a; chain: b; synonym: alpha-l-arabinanase, abn a, endo-1,5-alpha-l- arabinanase a; ec: 3.2.1.99; engineered: yes Organism, scientific name: Cellvibrio Japonicus; 1gyd contains a single unique chain 1gydB (315 residues long). 2 CHAIN 1GYDB 2.1 P95470 overview CONTENTS From SwissProt, id P95470, 99% identical to 1gydB: 1 Introduction 1 Description: Endo-a1,5-arabinanase (EC 3.2.1.99). Organism, scientific name: Cellvibrio japonicus. 2 Chain 1gydB 1 Taxonomy: Bacteria; Proteobacteria; Gammaproteobacteria; Pseu- 2.1 P95470 overview 1 domonadales; Pseudomonadaceae; Cellvibrio. 2.2 Multiple sequence alignment for 1gydB 1 2.3 Residue ranking in 1gydB 1 2.2 Multiple sequence alignment for 1gydB 2.4 Top ranking residues in 1gydB and their position on For the chain 1gydB, the alignment 1gydB.msf (attached) with 99 the structure 2 sequences was used. The alignment was downloaded from the HSSP 2.4.1 Clustering of residues at 25% coverage. 2 database, and fragments shorter than 75% of the query as well as 2.4.2 Possible novel functional surfaces at 25% duplicate sequences were removed. -

The Biology of Casmara Subagronoma (Lepidoptera
insects Article The Biology of Casmara subagronoma (Lepidoptera: Oecophoridae), a Stem-Boring Moth of Rhodomyrtus tomentosa (Myrtaceae): Descriptions of the Previously Unknown Adult Female and Immature Stages, and Its Potential as a Biological Control Candidate Susan A. Wineriter-Wright 1, Melissa C. Smith 1,* , Mark A. Metz 2 , Jeffrey R. Makinson 3 , Bradley T. Brown 3, Matthew F. Purcell 3, Kane L. Barr 4 and Paul D. Pratt 5 1 USDA-ARS Invasive Plant Research Laboratory, Fort Lauderdale, FL 33314, USA; [email protected] 2 USDA-ARS Systematic Entomology Lab, Beltsville, MD 20013-7012, USA; [email protected] 3 USDA-ARS Australian Biological Control Laboratory, CSIRO Health and Biosecurity, Dutton Park QLD 4102, Australia; jeff[email protected] (J.R.M.); [email protected] (B.T.B.); [email protected] (M.F.P.) 4 USDA-ARS Center for Medical, Agricultural and Veterinary Entomology, Gainesville, FL 32608, USA; [email protected] 5 USDA-ARS, Western Regional Research Center, Invasive Species and Pollinator Health Research Unit, 800 Buchanan Street, Albany, CA 94710, USA; [email protected] * Correspondence: [email protected]; Tel.: +1-954-475-6549 Received: 27 August 2020; Accepted: 16 September 2020; Published: 23 September 2020 Simple Summary: Rhodomyrtus tomentosa is a perennial woody shrub throughout Southeast Asia. Due to its prolific flower and fruit production, it was introduced into subtropical areas such as Florida and Hawai’i, where it is now naturalized and invasive. In an effort to find sustainable means to control R. tomentosa, a large-scale survey was mounted for biological control organisms. -

Acinetobacter Baumannii – a Neglected Pathogen in Veterinary and Environmental Health in Germany
Veterinary Research Communications (2019) 43:1–6 https://doi.org/10.1007/s11259-018-9742-0 REVIEW ARTICLE Acinetobacter baumannii – a neglected pathogen in veterinary and environmental health in Germany Gamal Wareth1 & Heinrich Neubauer1 & Lisa D. Sprague1 Received: 25 October 2018 /Accepted: 6 December 2018 /Published online: 27 December 2018 # The Author(s) 2018 Abstract The emergence and global spread of drug resistant Acinetobacter (A.) baumannii is a cause of great concern. The current knowledge on antibiotic resistance in A. baumannii from animal origin is mostly based on few internationally published case reports, investigations of strain collections and several whole genome analyses. This lack of data results in a somewhat sketchy picture on how to assess the possible impact of drug resistant A. baumannii strains on veterinary and public health in Germany. Consequently, there is an urgent need to intensify the surveillance of A. baumannii in pet animals, the farm animal population and wildlife. Keywords Acinetobacter baumannii . Review . Antibiotic resistance . Germany Introduction limited number of widespread clones appear to be responsible for hospital outbreaks in many countries (Diancourt et al. Acinetobacter (A.) baumannii is a gram-negative opportunis- 2010). Eight international clonal lineages have been described tic nosocomial pathogen belonging to the genus Acinetobacter so far and comparative typing of outbreak strains obtained all and a member of the family Moraxellaceae. Acinetobacter over Europe revealed the dominance of three clones, original- spp. are non-motile, non-fastidious Gram-negative, non- ly named European clones I-III (Dijkshoorn et al. 1996; Karah fermenting, catalase positive, oxidase negative, strictly aero- et al. -

Phylogenetic Analysis of Erwinia Species Based on 16S Rrna Gene Sequences?
INTERNATIONALJOURNAL OF SYSTEMATICBACTERIOLOGY, Oct. 1997, p. 1061-1067 Voi. 47, No. 4 0020-771 3/97/$04.00+0 Copyright 0 1997, International Union of Microbiological Societies Phylogenetic Analysis of Erwinia Species Based on 16s rRNA Gene Sequences? SOON-WO KWON,'" SEUNG-JOO GO,' HEE-WAN KANG,l JIN-CHANG RYU,l AND JIN-KI J02 National Institute of Agricultural Science & Technology, Suwon, and Department of Animal Science, Kyungpook National University, Daegu, Karea The phylogenetic relationships of the type strains of 16 Erwinia species were investigated by performing a comparative analysis of the sequences of the 16s rRNA genes of these organisms. The sequence data were analyzed by the neighbor-joining method, and each branch was supported by moderate bootstrap values. The phylogenetic tree and sequence analyses confirmed that the genus Erwinia is composed of species that exhibit considerable heterogeneity and form four clades that are intermixed with members of other genera, such as Escherichia coli, Klebsiella pneumoniae, and Serratia marcescens. Cluster I includes the type strains of Envinia herbicola, Erwinia milletiae, Erwinia ananas, Erwinia uredovora, and Erwinia stewartii and corresponds to Dye's herbicola group. Cluster I1 consists of Erwinia persicinus, Erwinia rhapontici, Erwinia amylovora, and Erwinia cypripedii. Cluster I11 consists of Erwinia carotovora subspecies and Erwinia chrysanthemi and is characterized by the production of pectate lyases and cellulases. Envinia salicis, Erwinia rubrifaciens, and Erwinia nigrijluens form the cluster that is most distantly related to other Erwinia species. The data from the sequence analyses are discussed in the context of biochemical and DNA-DNA hybridization data. The genus Erwinia was proposed by Winslow et al. -

Linking the Resistome and Plasmidome to the Microbiome
The ISME Journal (2019) 13:2437–2446 https://doi.org/10.1038/s41396-019-0446-4 ARTICLE Linking the resistome and plasmidome to the microbiome 1,2 3 3 3 1,2 Thibault Stalder ● Maximilian O. Press ● Shawn Sullivan ● Ivan Liachko ● Eva M. Top Received: 15 February 2019 / Revised: 2 May 2019 / Accepted: 10 May 2019 / Published online: 30 May 2019 © The Author(s) 2019. This article is published with open access Abstract The rapid spread of antibiotic resistance among bacterial pathogens is a serious human health threat. While a range of environments have been identified as reservoirs of antibiotic resistance genes (ARGs), we lack understanding of the origins of these ARGs and their spread from environment to clinic. This is partly due to our inability to identify the natural bacterial hosts of ARGs and the mobile genetic elements that mediate this spread, such as plasmids and integrons. Here we demonstrate that the in vivo proximity-ligation method Hi-C can reconstruct a known plasmid-host association from a wastewater community, and identify the in situ host range of ARGs, plasmids, and integrons by physically linking them to their host chromosomes. Hi-C detected both previously known and novel associations between ARGs, mobile genetic elements and host genomes, thus validating this method. We showed that IncQ plasmids and class 1 integrons had the broadest host range in this wastewater, and identified bacteria belonging to Moraxellaceae, Bacteroides,andPrevotella, and 1234567890();,: 1234567890();,: especially Aeromonadaceae as the most likely reservoirs of ARGs in this community. A better identification of the natural carriers of ARGs will aid the development of strategies to limit resistance spread to pathogens. -

Erwinia Chrysanthemi the Facts
Erwinia chrysanthemi (Dickeya spp.) The Facts Compiled for the British Potato Council by; Dr John Elphinstone, Central Science Laboratory Dr Ian Toth, Scottish Crop Research Institute Erwinia chrysanthemi (Dickeya spp.) – The Facts Contents Executive Summary 1. Introduction 2. The pathogen 3. Symptoms • Distinction from symptoms caused by P. atrosepticum and P. carotovorum • Distinction from other diseases 4. Geographical distribution • Presence in GB • Europe • Overseas 5. Biology, survival and dissemination of the pathogen • Factors influencing disease development • Dissemination • Survival 6. Assessment of Risk and Economic Loss • Quarantine status • Potential GB economic impact • Economic impact to overseas markets 7. Control • Statutory (certification) • On farm • Specific approaches and control measures in other countries • Best practice guide 8. Diagnostic methods 9. Knowledge gaps 10. Threats, Opportunities and Recommendations 11. References 12. Glossary of terms 2 © British Potato Council 2007 Erwinia chrysanthemi (Dickeya spp.) – The Facts Executive Summary The pathogen Erwinia chrysanthemi (Echr) is a complex of different bacteria now reclassified as species of Dickeya. While D. dadantii and D. zeae (formerly Echr biovar 3 or 8) are pathogens of potato in warmer countries, D. dianthicola (formerly Echr biovar 1 and 7) appears to be spreading on potatoes in Europe. The revised nomenclature of these pathogens has distinguished them from other soft rot erwiniae (including P. atrosepticum and P. carotovorum). Symptoms Symptoms of soft rot disease on potato tubers are similar whether caused by Dickeya or Pectobacterium spp. In the field, disease develops following movement of either pathogen from the stem base. Whereas P. atrosepticum typically causes blackleg symptoms under cool wet conditions, symptoms due to Dickeya spp. -

Characterization of Environmental and Cultivable Antibiotic- Resistant Microbial Communities Associated with Wastewater Treatment
antibiotics Article Characterization of Environmental and Cultivable Antibiotic- Resistant Microbial Communities Associated with Wastewater Treatment Alicia Sorgen 1, James Johnson 2, Kevin Lambirth 2, Sandra M. Clinton 3 , Molly Redmond 1 , Anthony Fodor 2 and Cynthia Gibas 2,* 1 Department of Biological Sciences, University of North Carolina at Charlotte, Charlotte, NC 28223, USA; [email protected] (A.S.); [email protected] (M.R.) 2 Department of Bioinformatics and Genomics, University of North Carolina at Charlotte, Charlotte, NC 28223, USA; [email protected] (J.J.); [email protected] (K.L.); [email protected] (A.F.) 3 Department of Geography & Earth Sciences, University of North Carolina at Charlotte, Charlotte, NC 28223, USA; [email protected] * Correspondence: [email protected]; Tel.: +1-704-687-8378 Abstract: Bacterial resistance to antibiotics is a growing global concern, threatening human and environmental health, particularly among urban populations. Wastewater treatment plants (WWTPs) are thought to be “hotspots” for antibiotic resistance dissemination. The conditions of WWTPs, in conjunction with the persistence of commonly used antibiotics, may favor the selection and transfer of resistance genes among bacterial populations. WWTPs provide an important ecological niche to examine the spread of antibiotic resistance. We used heterotrophic plate count methods to identify Citation: Sorgen, A.; Johnson, J.; phenotypically resistant cultivable portions of these bacterial communities and characterized the Lambirth, K.; Clinton, -

Genomic Encyclopedia of Sugar Utilization Pathways in The
Rodionov et al. BMC Genomics 2010, 11:494 http://www.biomedcentral.com/1471-2164/11/494 RESEARCH ARTICLE Open Access Genomic encyclopedia of sugar utilization pathways in the Shewanella genus Dmitry A Rodionov1,2, Chen Yang1,3, Xiaoqing Li1, Irina A Rodionova1, Yanbing Wang4, Anna Y Obraztsova4,7, Olga P Zagnitko5, Ross Overbeek5, Margaret F Romine6, Samantha Reed6, James K Fredrickson6, Kenneth H Nealson4,7, Andrei L Osterman1,5* Abstract Background: Carbohydrates are a primary source of carbon and energy for many bacteria. Accurate projection of known carbohydrate catabolic pathways across diverse bacteria with complete genomes constitutes a substantial challenge due to frequent variations in components of these pathways. To address a practically and fundamentally important challenge of reconstruction of carbohydrate utilization machinery in any microorganism directly from its genomic sequence, we combined a subsystems-based comparative genomic approach with experimental validation of selected bioinformatic predictions by a combination of biochemical, genetic and physiological experiments. Results: We applied this integrated approach to systematically map carbohydrate utilization pathways in 19 genomes from the Shewanella genus. The obtained genomic encyclopedia of sugar utilization includes ~170 protein families (mostly metabolic enzymes, transporters and transcriptional regulators) spanning 17 distinct pathways with a mosaic distribution across Shewanella species providing insights into their ecophysiology and adaptive evolution. Phenotypic -
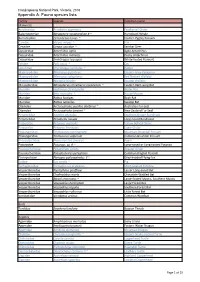
Report-VIC-Croajingolong National Park-Appendix A
Croajingolong National Park, Victoria, 2016 Appendix A: Fauna species lists Family Species Common name Mammals Acrobatidae Acrobates pygmaeus Feathertail Glider Balaenopteriae Megaptera novaeangliae # ~ Humpback Whale Burramyidae Cercartetus nanus ~ Eastern Pygmy Possum Canidae Vulpes vulpes ^ Fox Cervidae Cervus unicolor ^ Sambar Deer Dasyuridae Antechinus agilis Agile Antechinus Dasyuridae Antechinus mimetes Dusky Antechinus Dasyuridae Sminthopsis leucopus White-footed Dunnart Felidae Felis catus ^ Cat Leporidae Oryctolagus cuniculus ^ Rabbit Macropodidae Macropus giganteus Eastern Grey Kangaroo Macropodidae Macropus rufogriseus Red Necked Wallaby Macropodidae Wallabia bicolor Swamp Wallaby Miniopteridae Miniopterus schreibersii oceanensis ~ Eastern Bent-wing Bat Muridae Hydromys chrysogaster Water Rat Muridae Mus musculus ^ House Mouse Muridae Rattus fuscipes Bush Rat Muridae Rattus lutreolus Swamp Rat Otariidae Arctocephalus pusillus doriferus ~ Australian Fur-seal Otariidae Arctocephalus forsteri ~ New Zealand Fur Seal Peramelidae Isoodon obesulus Southern Brown Bandicoot Peramelidae Perameles nasuta Long-nosed Bandicoot Petauridae Petaurus australis Yellow Bellied Glider Petauridae Petaurus breviceps Sugar Glider Phalangeridae Trichosurus cunninghami Mountain Brushtail Possum Phalangeridae Trichosurus vulpecula Common Brushtail Possum Phascolarctidae Phascolarctos cinereus Koala Potoroidae Potorous sp. # ~ Long-nosed or Long-footed Potoroo Pseudocheiridae Petauroides volans Greater Glider Pseudocheiridae Pseudocheirus peregrinus -

Ultradeep Microbial Communities at 4.4 Km Within Crystalline Bedrock: Implications for Habitability in a Planetary Context
life Article Ultradeep Microbial Communities at 4.4 km within Crystalline Bedrock: Implications for Habitability in a Planetary Context Lotta Purkamo 1,2,* , Riikka Kietäväinen 2,3, Maija Nuppunen-Puputti 4, Malin Bomberg 4 and Claire Cousins 1 1 School of Earth and Environmental Sciences, University of St Andrews, St Andrews KY16 9AL, UK; [email protected] 2 Geological Survey of Finland, 02151 Espoo, Finland; riikka.kietavainen@gtk.fi 3 Department of Geosciences and Geography, University of Helsinki, 00014 Helsinki, Finland 4 VTT Technical Research Centre of Finland, 02044 VTT, Finland; maija.nuppunen-puputti@vtt.fi (M.N.-P.); malin.bomberg@vtt.fi (M.B.) * Correspondence: lotta.purkamo@gtk.fi; Tel.: +358-44-322-9432 Received: 1 November 2019; Accepted: 1 January 2020; Published: 4 January 2020 Abstract: The deep bedrock surroundings are an analog for extraterrestrial habitats for life. In this study, we investigated microbial life within anoxic ultradeep boreholes in Precambrian bedrock, including the adaptation to environmental conditions and lifestyle of these organisms. Samples were collected from Pyhäsalmi mine environment in central Finland and from geothermal drilling wells in Otaniemi, Espoo, in southern Finland. Microbial communities inhabiting the up to 4.4 km deep bedrock were characterized with phylogenetic marker gene (16S rRNA genes and fungal ITS region) amplicon and DNA and cDNA metagenomic sequencing. Functional marker genes (dsrB, mcrA, narG) were quantified with qPCR. Results showed that although crystalline bedrock provides very limited substrates for life, the microbial communities are diverse. Gammaproteobacterial phylotypes were most dominant in both studied sites. Alkanindiges -affiliating OTU was dominating in Pyhäsalmi fluids, while different depths of Otaniemi samples were dominated by Pseudomonas. -

Data Sheet on Erwinia Chrysanthemi
EPPO quarantine pest Prepared by CABI and EPPO for the EU under Contract 90/399003 Data Sheets on Quarantine Pests Erwinia chrysanthemi IDENTITY Name: Erwinia chrysanthemi Burkholder et al. Synonyms: Erwinia carotovora (Jones) Bergey et al. f.sp. parthenii Starr Erwinia carotovora (Jones) Bergey et al. f.sp. dianthicola (Hellmers) Bakker Pectobacterium parthenii (Starr) Hellmers Erwinia carotovora (Jones) Bergey et al. var. chrysanthemi (Burkholder et al.) Dye Taxonomic position: Bacteria: Gracilicutes Notes on taxonomy and nomenclature: Six pathovars of E. chrysanthemi are mentioned in the Bergey's Manual of Systematic Bacteriology (pv. chrysanthemi, pv. dianthicola, pv. dieffenbachiae, pv. paradisiaca, pv. parthenii and pv. zeae) on the basis of their host plants (Lelliott & Dickey, 1984). The pathovars are more or less related to six biochemical subdivisions (I-VI, Dickey & Victoria, 1980). Nine biovars (1-9; Ngwira & Samson, 1990) have been proposed for non-ambiguous biochemical typing. Some of the biochemical variants may be elevated to subspecies level or to another species name in the near future. Bayer computer code: ERWICH (also ERWIZE) EPPO A2 list: No. 53 EU Annex designation: II/A2 - pv. dianthicola only HOSTS Diseases have most often been reported on bananas, carnations, chrysanthemums, Dahlia, Dieffenbachia spp., Euphorbia pulcherrima, Kalanchoe blossfeldiana, maize, Philodendron spp., potatoes, Saintpaulia ionantha, Syngonium podophyllum. E. chrysanthemi has also been naturally found to attack Allium fistulosum, Brassica chinensis, Capsicum, cardamoms, carrots, celery, chicory, Colocasia esculenta, Poaceae such as Brachiaria mutica, B. ruziziensis, Panicum maximum and Pennisetum purpureum, Hyacinthus sp., Leucanthemum maximum, lucerne, onions, pineapples, radishes, rice, Sedum spectabile, sugarcane, sorghum, sweet potatoes, tobacco, tomatoes, tulips and glasshouse ornamentals such as Aechmea fasciata, Aglaonema pictum, Anemone spp., Begonia intermedia cv.