Identification and Characterization of Two Novel Syncytin-Like Retroviral Envelope Genes, Captured for a Possible Role in the At
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Reproductive Specializations in a Viviparous African Skink: Implications for Evolution and Biological Conservation Daniel G
Trinity College Trinity College Digital Repository Faculty Scholarship 8-2010 Reproductive Specializations in a Viviparous African Skink: Implications for Evolution and Biological Conservation Daniel G. Blackburn Trinity College, [email protected] Alexander F. Flemming University of Stellenbosch Follow this and additional works at: http://digitalrepository.trincoll.edu/facpub Part of the Biology Commons Herpetological Conservation and Biology 5(2):263-270. Symposium: Reptile Reproduction REPRODUCTIVE SPECIALIZATIONS IN A VIVIPAROUS AFRICAN SKINK AND ITS IMPLICATIONS FOR EVOLUTION AND CONSERVATION 1 2 DANIEL G. BLACKBURN AND ALEXANDER F. FLEMMING 1Department of Biology and Electron Microscopy Facility, Trinity College, Hartford, Connecticut 06106, USA, e-mail: [email protected] 2Department of Botany and Zoology, University of Stellenbosch, Stellenbosch 7600, South Africa Abstract.—Recent research on the African scincid lizard, Trachylepis ivensi, has significantly expanded the range of known reproductive specializations in reptiles. This species is viviparous and exhibits characteristics previously thought to be confined to therian mammals. In most viviparous squamates, females ovulate large yolk-rich eggs that provide most of the nutrients for development. Typically, their placental components (fetal membranes and uterus) are relatively unspecialized, and similar to their oviparous counterparts. In T. ivensi, females ovulate tiny eggs and provide nutrients for embryonic development almost entirely by placental means. Early in gestation, embryonic tissues invade deeply into maternal tissues and establish an intimate “endotheliochorial” relationship with the maternal blood supply by means of a yolk sac placenta. The presence of such an invasive form of implantation in a squamate reptile is unprecedented and has significant functional and evolutionary implications. Discovery of the specializations of T. -

The Herpetofauna of the Cubango, Cuito, and Lower Cuando River Catchments of South-Eastern Angola
Official journal website: Amphibian & Reptile Conservation amphibian-reptile-conservation.org 10(2) [Special Section]: 6–36 (e126). The herpetofauna of the Cubango, Cuito, and lower Cuando river catchments of south-eastern Angola 1,2,*Werner Conradie, 2Roger Bills, and 1,3William R. Branch 1Port Elizabeth Museum (Bayworld), P.O. Box 13147, Humewood 6013, SOUTH AFRICA 2South African Institute for Aquatic Bio- diversity, P/Bag 1015, Grahamstown 6140, SOUTH AFRICA 3Research Associate, Department of Zoology, P O Box 77000, Nelson Mandela Metropolitan University, Port Elizabeth 6031, SOUTH AFRICA Abstract.—Angola’s herpetofauna has been neglected for many years, but recent surveys have revealed unknown diversity and a consequent increase in the number of species recorded for the country. Most historical Angola surveys focused on the north-eastern and south-western parts of the country, with the south-east, now comprising the Kuando-Kubango Province, neglected. To address this gap a series of rapid biodiversity surveys of the upper Cubango-Okavango basin were conducted from 2012‒2015. This report presents the results of these surveys, together with a herpetological checklist of current and historical records for the Angolan drainage of the Cubango, Cuito, and Cuando Rivers. In summary 111 species are known from the region, comprising 38 snakes, 32 lizards, five chelonians, a single crocodile and 34 amphibians. The Cubango is the most western catchment and has the greatest herpetofaunal diversity (54 species). This is a reflection of both its easier access, and thus greatest number of historical records, and also the greater habitat and topographical diversity associated with the rocky headwaters. -

Arrival and Diversification of Mabuyine Skinks (Squamata: Scincidae) in the Neotropics Based on a Fossil-Calibrated Timetree
Arrival and diversification of mabuyine skinks (Squamata: Scincidae) in the Neotropics based on a fossil-calibrated timetree Anieli Guirro Pereira and Carlos G. Schrago Department of Genetics, Federal University of Rio de Janeiro, Rio de Janeiro, RJ, Brazil ABSTRACT Background. The evolution of South American Mabuyinae skinks holds significant biogeographic interest because its sister lineage is distributed across the African continent and adjacent islands. Moreover, at least one insular species, Trachylepis atlantica, has independently reached the New World through transoceanic dispersal. To clarify the evolutionary history of both Neotropical lineages, this study aimed to infer an updated timescale using the largest species and gene sampling dataset ever assembled for this group. By extending the analysis to the Scincidae family, we could employ fossil information to estimate mabuyinae divergence times and carried out a formal statistical biogeography analysis. To unveil macroevolutionary patterns, we also inferred diversification rates for this lineage and evaluated whether the colonization of South American continent significantly altered the mode of Mabuyinae evolution. Methods. A time-calibrated phylogeny was inferred under the Bayesian framework employing fossil information. This timetree was used to (i) evaluate the historical biogeography of mabuiyines using the statistical approach implemented in Bio- GeoBEARS; (ii) estimate macroevolutionary diversification rates of the South American Mabuyinae lineages and the patterns of evolution of selected traits, namely, the mode of reproduction, body mass and snout–vent length; (iii) test the hypothesis of differential macroevolutionary patterns in South American lineages in BAMM and GeoSSE; and Submitted 21 November 2016 (iv) re-evaluate the ancestral state of the mode of reproduction of mabuyines. -

Reproductionreview
REPRODUCTIONREVIEW The evolution of viviparity: molecular and genomic data from squamate reptiles advance understanding of live birth in amniotes James U Van Dyke, Matthew C Brandley and Michael B Thompson School of Biological Sciences, University of Sydney, A08 Heydon-Laurence Building, Sydney, New South Wales 2006, Australia Correspondence should be addressed to J U Van Dyke; Email: [email protected] Abstract Squamate reptiles (lizards and snakes) are an ideal model system for testing hypotheses regarding the evolution of viviparity (live birth) in amniote vertebrates. Viviparity has evolved over 100 times in squamates, resulting in major changes in reproductive physiology. At a minimum, all viviparous squamates exhibit placentae formed by the appositions of maternal and embryonic tissues, which are homologous in origin with the tissues that form the placenta in therian mammals. These placentae facilitate adhesion of the conceptus to the uterus as well as exchange of oxygen, carbon dioxide, water, sodium, and calcium. However, most viviparous squamates continue to rely on yolk for nearly all of their organic nutrition. In contrast, some species, which rely on the placenta for at least a portion of organic nutrition, exhibit complex placental specializations associated with the transport of amino acids and fatty acids. Some viviparous squamates also exhibit reduced immunocompetence during pregnancy, which could be the result of immunosuppression to protect developing embryos. Recent molecular studies using both candidate-gene and next-generation sequencing approaches have suggested that at least some of the genes and gene families underlying these phenomena play similar roles in the uterus and placenta of viviparous mammals and squamates. -

An Endogenous Retroviral Envelope Syncytin and Its Cognate Receptor Identified in the Viviparous Placental Mabuya Lizard
An endogenous retroviral envelope syncytin and its PNAS PLUS cognate receptor identified in the viviparous placental Mabuya lizard Guillaume Cornelisa,b,1,2, Mathis Funka,b,1, Cécile Vernocheta,b, Francisca Lealc,3, Oscar Alejandro Tarazonac,4, Guillaume Meuriced, Odile Heidmanna,b, Anne Dupressoira,b, Aurélien Mirallese, Martha Patricia Ramirez-Pinillac, and Thierry Heidmanna,b,5 SEE COMMENTARY aUnité Physiologie et Pathologie Moléculaires des Rétrovirus Endogènes et Infectieux, CNRS UMR 9196, Gustave Roussy, Villejuif, F-94805, France; bUMR 9196, Université Paris-Sud, Orsay, F-91405, France; cLaboratorio de Biologia Reproductiva de Vertebrados, Escuela de Biologia, Universidad Industrial de Santander, 680002 Bucaramanga, Colombia; dPlateforme de Bioinformatique, INSERM US23/CNRS UMS3655, Gustave Roussy, Villejuif, F-94805, France; and eInstitut de Systématique, Evolution, Biodiversité, Muséum National d’Histoire Naturelle, CNRS UPMC EPHE, Sorbonne Universités, Paris, F-75005, France Edited by R. Michael Roberts, University of Missouri-Columbia, Columbia, MO, and approved October 26, 2017 (received for review August 23, 2017) Syncytins are envelope genes from endogenous retroviruses that Remarkably, placental structures are not restricted to mamma- have been captured during evolution for a function in placentation. lian species. Placentation emerged independently and in a sto- They have been found in all placental mammals in which they have chastic manner in several groups of vertebrates, with the noticeable been searched, including marsupials. Placental structures are not exception of birds (reviewed in refs. 16 and 17). In particular, restricted to mammals but also emerged in some other vertebrates, complex placentas have been described in some South American most frequently in lizards, such as the viviparous Mabuya Scincidae. -

Reproductive Specializations in a Viviparous African Skink: Implications for Evolution and Biological Conservation
Trinity College Trinity College Digital Repository Faculty Scholarship 8-2010 Reproductive Specializations in a Viviparous African Skink: Implications for Evolution and Biological Conservation Daniel G. Blackburn Trinity College, [email protected] Alexander F. Flemming University of Stellenbosch Follow this and additional works at: https://digitalrepository.trincoll.edu/facpub Part of the Biology Commons Herpetological Conservation and Biology 5(2):263-270. Symposium: Reptile Reproduction REPRODUCTIVE SPECIALIZATIONS IN A VIVIPAROUS AFRICAN SKINK AND ITS IMPLICATIONS FOR EVOLUTION AND CONSERVATION 1 2 DANIEL G. BLACKBURN AND ALEXANDER F. FLEMMING 1Department of Biology and Electron Microscopy Facility, Trinity College, Hartford, Connecticut 06106, USA, e-mail: [email protected] 2Department of Botany and Zoology, University of Stellenbosch, Stellenbosch 7600, South Africa Abstract.—Recent research on the African scincid lizard, Trachylepis ivensi, has significantly expanded the range of known reproductive specializations in reptiles. This species is viviparous and exhibits characteristics previously thought to be confined to therian mammals. In most viviparous squamates, females ovulate large yolk-rich eggs that provide most of the nutrients for development. Typically, their placental components (fetal membranes and uterus) are relatively unspecialized, and similar to their oviparous counterparts. In T. ivensi, females ovulate tiny eggs and provide nutrients for embryonic development almost entirely by placental means. Early in gestation, embryonic tissues invade deeply into maternal tissues and establish an intimate “endotheliochorial” relationship with the maternal blood supply by means of a yolk sac placenta. The presence of such an invasive form of implantation in a squamate reptile is unprecedented and has significant functional and evolutionary implications. Discovery of the specializations of T. -

Patterns of Species Richness, Endemism and Environmental Gradients of African Reptiles
Journal of Biogeography (J. Biogeogr.) (2016) ORIGINAL Patterns of species richness, endemism ARTICLE and environmental gradients of African reptiles Amir Lewin1*, Anat Feldman1, Aaron M. Bauer2, Jonathan Belmaker1, Donald G. Broadley3†, Laurent Chirio4, Yuval Itescu1, Matthew LeBreton5, Erez Maza1, Danny Meirte6, Zoltan T. Nagy7, Maria Novosolov1, Uri Roll8, 1 9 1 1 Oliver Tallowin , Jean-Francßois Trape , Enav Vidan and Shai Meiri 1Department of Zoology, Tel Aviv University, ABSTRACT 6997801 Tel Aviv, Israel, 2Department of Aim To map and assess the richness patterns of reptiles (and included groups: Biology, Villanova University, Villanova PA 3 amphisbaenians, crocodiles, lizards, snakes and turtles) in Africa, quantify the 19085, USA, Natural History Museum of Zimbabwe, PO Box 240, Bulawayo, overlap in species richness of reptiles (and included groups) with the other ter- Zimbabwe, 4Museum National d’Histoire restrial vertebrate classes, investigate the environmental correlates underlying Naturelle, Department Systematique et these patterns, and evaluate the role of range size on richness patterns. Evolution (Reptiles), ISYEB (Institut Location Africa. Systematique, Evolution, Biodiversite, UMR 7205 CNRS/EPHE/MNHN), Paris, France, Methods We assembled a data set of distributions of all African reptile spe- 5Mosaic, (Environment, Health, Data, cies. We tested the spatial congruence of reptile richness with that of amphib- Technology), BP 35322 Yaounde, Cameroon, ians, birds and mammals. We further tested the relative importance of 6Department of African Biology, Royal temperature, precipitation, elevation range and net primary productivity for Museum for Central Africa, 3080 Tervuren, species richness over two spatial scales (ecoregions and 1° grids). We arranged Belgium, 7Royal Belgian Institute of Natural reptile and vertebrate groups into range-size quartiles in order to evaluate the Sciences, OD Taxonomy and Phylogeny, role of range size in producing richness patterns. -

Reptiles & Amphibians
AWF FOUR CORNERS TBNRM PROJECT : REVIEWS OF EXISTING BIODIVERSITY INFORMATION i Published for The African Wildlife Foundation's FOUR CORNERS TBNRM PROJECT by THE ZAMBEZI SOCIETY and THE BIODIVERSITY FOUNDATION FOR AFRICA 2004 PARTNERS IN BIODIVERSITY The Zambezi Society The Biodiversity Foundation for Africa P O Box HG774 P O Box FM730 Highlands Famona Harare Bulawayo Zimbabwe Zimbabwe Tel: +263 4 747002-5 E-mail: [email protected] E-mail: [email protected] Website: www.biodiversityfoundation.org Website : www.zamsoc.org The Zambezi Society and The Biodiversity Foundation for Africa are working as partners within the African Wildlife Foundation's Four Corners TBNRM project. The Biodiversity Foundation for Africa is responsible for acquiring technical information on the biodiversity of the project area. The Zambezi Society will be interpreting this information into user-friendly formats for stakeholders in the Four Corners area, and then disseminating it to these stakeholders. THE BIODIVERSITY FOUNDATION FOR AFRICA (BFA is a non-profit making Trust, formed in Bulawayo in 1992 by a group of concerned scientists and environmentalists. Individual BFA members have expertise in biological groups including plants, vegetation, mammals, birds, reptiles, fish, insects, aquatic invertebrates and ecosystems. The major objective of the BFA is to undertake biological research into the biodiversity of sub-Saharan Africa, and to make the resulting information more accessible. Towards this end it provides technical, ecological and biosystematic expertise. THE ZAMBEZI SOCIETY was established in 1982. Its goals include the conservation of biological diversity and wilderness in the Zambezi Basin through the application of sustainable, scientifically sound natural resource management strategies. -
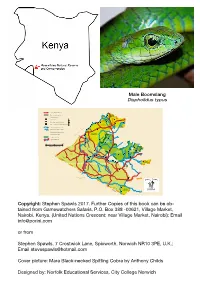
Guide to the Reptiles and Amphibians of the Maasai Mara
Male Boomslang Dispholidus typus N OLARE MOTOROGI CONSERVANCY OLARRO CONSERVANCY Copyright: Stephen Spawls 2017. Further Copies of this book can be ob- tained from Gamewatchers Safaris, P.O. Box 388 -00621, Village Market, Nairobi, Kenya, (United Nations Crescent; near Village Market, Nairobi); Email [email protected] or from Stephen Spawls, 7 Crostwick Lane, Spixworth, Norwich NR10 3PE, U.K.; Email [email protected] Cover picture: Mara Black-necked Spitting Cobra by Anthony Childs Designed by: Norfolk Educational Services, City College Norwich A Guide to Amphibians and Reptiles of the Maasai Mara Ecosystem By Printed by Stephen Spawls Norwich City College Print Room Contents Introduction 2 Foreword by Jake Grieves-Cook, 3 Chairman of Gamewatchers Safaris & Porini Camps Why conservation and the conservancies are important 4 Observing and Collecting Reptiles and Amphibians 5 Safety and Reptiles/Amphibians 7 Snakebite 8 The Maasai-Mara Ecosystem and its reptile 9 and amphibian fauna The Reptiles and Amphibians of the Maasai Mara ecosystem 10 Some species that might be found in the Maasai Mara 34 but are as yet unrecorded Acknowledgements 38 Further Resources 39 Introduction “The natural resources of this country – its wildlife which offers such an attraction to beautiful places…the mighty forests which guard the water catchment areas so vital to the survival of man and beast – are a priceless heritage for the future. The government of Kenya…pledges itself to conserve them for posterity with all the means at its disposal.” Mzee Jomo Kenyatta, First President of the Republic of Kenya (inscription at the entrance of Nairobi National Park 1964) As far as I am aware, this book is the first guide to the reptiles and amphibians of any large conservation area in Kenya. -

DNA Barcoding Assessment of Genetic Variation in Two Widespread Skinks from Madagascar, Trachylepis Elegans and T
Zootaxa 3755 (5): 477–484 ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ Article ZOOTAXA Copyright © 2014 Magnolia Press ISSN 1175-5334 (online edition) http://dx.doi.org/10.11646/zootaxa.3755.5.7 http://zoobank.org/urn:lsid:zoobank.org:pub:039C3026-D3B9-47B2-8340-6B916C636665 DNA barcoding assessment of genetic variation in two widespread skinks from Madagascar, Trachylepis elegans and T. gravenhorstii (Squamata: Scincidae) MIGUEL VENCES1,6*, ALEXANDRA LIMA2,3*, AURÉLIEN MIRALLES4 & FRANK GLAW5 1Zoological Institute, Technische Universität Braunschweig, Mendelssohnstr. 4, 38106 Braunschweig, Germany 2Centro de Investigação em Biodiversidade e Recursos Genéticos (CIBIO), Campus Agrário de Vairão, R. Padre Armando Quintas, 4485-661 Vairão, Portugal 3Departamento de Biologia, Faculdade de Ciências, Universidade do Porto, R. Campo Alegre s/n, 4169-007 Porto, Portugal 4CNRS-UMR5175 Centre d'Ecologie Fonctionnelle et Evolutive, 1919 route de Mende, 34293 Montpellier cedex 5, France 5Zoologische Staatssammlung München, Münchhausenstr. 21, 81247 München, Germany 6Corresponding author. E-mail: [email protected] *These authors contributed equally to the paper Abstract Trachylepis elegans and T. gravenhorstii are two of the most widespread reptiles in Madagascar, inhabiting a wide variety of habitats. Previous studies have indicated a considerable mitochondrial DNA (mtDNA) variation within these species, but the geographic distribution of the major haplotype lineages is poorly known. Herein we analyse the phylogeography of these lizards based on 107 sequences of the mitochondrial cytochrome oxidase subunit I gene, 101 of which newly de- termined. As in previous mtDNA assessments, T. elegans and T. gravenhorstii were not reciprocally monophyletic, al- though recent analyses including nuclear markers indicated their probable monophyly, respectively. -

An Endogenous Retroviral Envelope Syncytin and Its Cognate Receptor
An endogenous retroviral envelope syncytin and its PNAS PLUS cognate receptor identified in the viviparous placental Mabuya lizard Guillaume Cornelisa,b,1,2, Mathis Funka,b,1, Cécile Vernocheta,b, Francisca Lealc,3, Oscar Alejandro Tarazonac,4, Guillaume Meuriced, Odile Heidmanna,b, Anne Dupressoira,b, Aurélien Mirallese, Martha Patricia Ramirez-Pinillac, and Thierry Heidmanna,b,5 aUnité Physiologie et Pathologie Moléculaires des Rétrovirus Endogènes et Infectieux, CNRS UMR 9196, Gustave Roussy, Villejuif, F-94805, France; bUMR 9196, Université Paris-Sud, Orsay, F-91405, France; cLaboratorio de Biologia Reproductiva de Vertebrados, Escuela de Biologia, Universidad Industrial de Santander, 680002 Bucaramanga, Colombia; dPlateforme de Bioinformatique, INSERM US23/CNRS UMS3655, Gustave Roussy, Villejuif, F-94805, France; and eInstitut de Systématique, Evolution, Biodiversité, Muséum National d’Histoire Naturelle, CNRS UPMC EPHE, Sorbonne Universités, Paris, F-75005, France Edited by R. Michael Roberts, University of Missouri-Columbia, Columbia, MO, and approved October 26, 2017 (received for review August 23, 2017) Syncytins are envelope genes from endogenous retroviruses that Remarkably, placental structures are not restricted to mamma- have been captured during evolution for a function in placentation. lian species. Placentation emerged independently and in a sto- They have been found in all placental mammals in which they have chastic manner in several groups of vertebrates, with the noticeable been searched, including marsupials. Placental structures are not exception of birds (reviewed in refs. 16 and 17). In particular, restricted to mammals but also emerged in some other vertebrates, complex placentas have been described in some South American most frequently in lizards, such as the viviparous Mabuya Scincidae. -

Here Additional S�Ecies E��Ected in �Undavala �Ere Recorded
© University of Hamburg 2018 All rights reserved Klaus Hess Publishers Göttingen & Windhoek www.k-hess-verlag.de ISBN: 978-3-933117-95-3 (Germany), 978-99916-57-43-1 (Namibia) Language editing: Will Simonson (Cambridge), and Proofreading Pal Translation of abstracts to Portuguese: Ana Filipa Guerra Silva Gomes da Piedade Page desing & layout: Marit Arnold, Klaus A. Hess, Ria Henning-Lohmann Cover photographs: front: Thunderstorm approaching a village on the Angolan Central Plateau (Rasmus Revermann) back: Fire in the miombo woodlands, Zambia (David Parduhn) Cover Design: Ria Henning-Lohmann ISSN 1613-9801 Printed in Germany Suggestion for citations: Volume: Revermann, R., Krewenka, K.M., Schmiedel, U., Olwoch, J.M., Helmschrot, J. & Jürgens, N. (eds.) (2018) Climate change and adaptive land management in southern Africa – assessments, changes, challenges, and solutions. Biodiversity & Ecology, 6, Klaus Hess Publishers, Göttingen & Windhoek. Articles (example): Archer, E., Engelbrecht, F., Hänsler, A., Landman, W., Tadross, M. & Helmschrot, J. (2018) Seasonal prediction and regional climate projections for southern Africa. In: Climate change and adaptive land management in southern Africa – assessments, changes, challenges, and solutions (ed. by Revermann, R., Krewenka, K.M., Schmiedel, U., Olwoch, J.M., Helmschrot, J. & Jürgens, N.), pp. 14–21, Biodiversity & Ecology, 6, Klaus Hess Publishers, Göttingen & Windhoek. Corrections brought to our attention will be published at the following location: http://www.biodiversity-plants.de/biodivers_ecol/biodivers_ecol.php