Thèse De Doctorat
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Method to Infer Changed Activity of Metabolic Function from Transcript Profiles
ModeScore: A Method to Infer Changed Activity of Metabolic Function from Transcript Profiles Andreas Hoppe and Hermann-Georg Holzhütter Charité University Medicine Berlin, Institute for Biochemistry, Computational Systems Biochemistry Group [email protected] Abstract Genome-wide transcript profiles are often the only available quantitative data for a particular perturbation of a cellular system and their interpretation with respect to the metabolism is a major challenge in systems biology, especially beyond on/off distinction of genes. We present a method that predicts activity changes of metabolic functions by scoring reference flux distributions based on relative transcript profiles, providing a ranked list of most regulated functions. Then, for each metabolic function, the involved genes are ranked upon how much they represent a specific regulation pattern. Compared with the naïve pathway-based approach, the reference modes can be chosen freely, and they represent full metabolic functions, thus, directly provide testable hypotheses for the metabolic study. In conclusion, the novel method provides promising functions for subsequent experimental elucidation together with outstanding associated genes, solely based on transcript profiles. 1998 ACM Subject Classification J.3 Life and Medical Sciences Keywords and phrases Metabolic network, expression profile, metabolic function Digital Object Identifier 10.4230/OASIcs.GCB.2012.1 1 Background The comprehensive study of the cell’s metabolism would include measuring metabolite concentrations, reaction fluxes, and enzyme activities on a large scale. Measuring fluxes is the most difficult part in this, for a recent assessment of techniques, see [31]. Although mass spectrometry allows to assess metabolite concentrations in a more comprehensive way, the larger the set of potential metabolites, the more difficult [8]. -

The Kyoto Encyclopedia of Genes and Genomes (KEGG)
Kyoto Encyclopedia of Genes and Genome Minoru Kanehisa Institute for Chemical Research, Kyoto University HFSPO Workshop, Strasbourg, November 18, 2016 The KEGG Databases Category Database Content PATHWAY KEGG pathway maps Systems information BRITE BRITE functional hierarchies MODULE KEGG modules KO (KEGG ORTHOLOGY) KO groups for functional orthologs Genomic information GENOME KEGG organisms, viruses and addendum GENES / SSDB Genes and proteins / sequence similarity COMPOUND Chemical compounds GLYCAN Glycans Chemical information REACTION / RCLASS Reactions / reaction classes ENZYME Enzyme nomenclature DISEASE Human diseases DRUG / DGROUP Drugs / drug groups Health information ENVIRON Health-related substances (KEGG MEDICUS) JAPIC Japanese drug labels DailyMed FDA drug labels 12 manually curated original DBs 3 DBs taken from outside sources and given original annotations (GENOME, GENES, ENZYME) 1 computationally generated DB (SSDB) 2 outside DBs (JAPIC, DailyMed) KEGG is widely used for functional interpretation and practical application of genome sequences and other high-throughput data KO PATHWAY GENOME BRITE DISEASE GENES MODULE DRUG Genome Molecular High-level Practical Metagenome functions functions applications Transcriptome etc. Metabolome Glycome etc. COMPOUND GLYCAN REACTION Funding Annual budget Period Funding source (USD) 1995-2010 Supported by 10+ grants from Ministry of Education, >2 M Japan Society for Promotion of Science (JSPS) and Japan Science and Technology Agency (JST) 2011-2013 Supported by National Bioscience Database Center 0.8 M (NBDC) of JST 2014-2016 Supported by NBDC 0.5 M 2017- ? 1995 KEGG website made freely available 1997 KEGG FTP site made freely available 2011 Plea to support KEGG KEGG FTP academic subscription introduced 1998 First commercial licensing Contingency Plan 1999 Pathway Solutions Inc. -

3 13437143.Pdf
Title Integrative Annotation of 21,037 Human Genes Validated by Full-Length cDNA Clones Imanishi, Tadashi; Itoh, Takeshi; Suzuki, Yutaka; O'Donovan, Claire; Fukuchi, Satoshi; Koyanagi, Kanako O.; Barrero, Roberto A.; Tamura, Takuro; Yamaguchi-Kabata, Yumi; Tanino, Motohiko; Yura, Kei; Miyazaki, Satoru; Ikeo, Kazuho; Homma, Keiichi; Kasprzyk, Arek; Nishikawa, Tetsuo; Hirakawa, Mika; Thierry-Mieg, Jean; Thierry-Mieg, Danielle; Ashurst, Jennifer; Jia, Libin; Nakao, Mitsuteru; Thomas, Michael A.; Mulder, Nicola; Karavidopoulou, Youla; Jin, Lihua; Kim, Sangsoo; Yasuda, Tomohiro; Lenhard, Boris; Eveno, Eric; Suzuki, Yoshiyuki; Yamasaki, Chisato; Takeda, Jun-ichi; Gough, Craig; Hilton, Phillip; Fujii, Yasuyuki; Sakai, Hiroaki; Tanaka, Susumu; Amid, Clara; Bellgard, Matthew; Bonaldo, Maria de Fatima; Bono, Hidemasa; Bromberg, Susan K.; Brookes, Anthony J.; Bruford, Elspeth; Carninci, Piero; Chelala, Claude; Couillault, Christine; Souza, Sandro J. de; Debily, Marie-Anne; Devignes, Marie-Dominique; Dubchak, Inna; Endo, Toshinori; Estreicher, Anne; Eyras, Eduardo; Fukami-Kobayashi, Kaoru; R. Gopinath, Gopal; Graudens, Esther; Hahn, Yoonsoo; Han, Michael; Han, Ze-Guang; Hanada, Kousuke; Hanaoka, Hideki; Harada, Erimi; Hashimoto, Katsuyuki; Hinz, Ursula; Hirai, Momoki; Hishiki, Teruyoshi; Hopkinson, Ian; Imbeaud, Sandrine; Inoko, Hidetoshi; Kanapin, Alexander; Kaneko, Yayoi; Kasukawa, Takeya; Kelso, Janet; Kersey, Author(s) Paul; Kikuno, Reiko; Kimura, Kouichi; Korn, Bernhard; Kuryshev, Vladimir; Makalowska, Izabela; Makino, Takashi; Mano, Shuhei; -

The Principled Design of Large-Scale Recursive Neural Network Architectures–DAG-Rnns and the Protein Structure Prediction Problem
Journal of Machine Learning Research 4 (2003) 575-602 Submitted 2/02; Revised 4/03; Published 9/03 The Principled Design of Large-Scale Recursive Neural Network Architectures–DAG-RNNs and the Protein Structure Prediction Problem Pierre Baldi [email protected] Gianluca Pollastri [email protected] School of Information and Computer Science Institute for Genomics and Bioinformatics University of California, Irvine Irvine, CA 92697-3425, USA Editor: Michael I. Jordan Abstract We describe a general methodology for the design of large-scale recursive neural network architec- tures (DAG-RNNs) which comprises three fundamental steps: (1) representation of a given domain using suitable directed acyclic graphs (DAGs) to connect visible and hidden node variables; (2) parameterization of the relationship between each variable and its parent variables by feedforward neural networks; and (3) application of weight-sharing within appropriate subsets of DAG connec- tions to capture stationarity and control model complexity. Here we use these principles to derive several specific classes of DAG-RNN architectures based on lattices, trees, and other structured graphs. These architectures can process a wide range of data structures with variable sizes and dimensions. While the overall resulting models remain probabilistic, the internal deterministic dy- namics allows efficient propagation of information, as well as training by gradient descent, in order to tackle large-scale problems. These methods are used here to derive state-of-the-art predictors for protein structural features such as secondary structure (1D) and both fine- and coarse-grained contact maps (2D). Extensions, relationships to graphical models, and implications for the design of neural architectures are briefly discussed. -

Methodology for Predicting Semantic Annotations of Protein Sequences by Feature Extraction Derived of Statistical Contact Potentials and Continuous Wavelet Transform
Universidad Nacional de Colombia Sede Manizales Master’s Thesis Methodology for predicting semantic annotations of protein sequences by feature extraction derived of statistical contact potentials and continuous wavelet transform Author: Supervisor: Gustavo Alonso Arango Dr. Cesar German Argoty Castellanos Dominguez A thesis submitted in fulfillment of the requirements for the degree of Master’s on Engineering - Industrial Automation in the Department of Electronic, Electric Engineering and Computation Signal Processing and Recognition Group June 2014 Universidad Nacional de Colombia Sede Manizales Tesis de Maestr´ıa Metodolog´ıapara predecir la anotaci´on sem´antica de prote´ınaspor medio de extracci´on de caracter´ısticas derivadas de potenciales de contacto y transformada wavelet continua Autor: Tutor: Gustavo Alonso Arango Dr. Cesar German Argoty Castellanos Dominguez Tesis presentada en cumplimiento a los requerimientos necesarios para obtener el grado de Maestr´ıaen Ingenier´ıaen Automatizaci´onIndustrial en el Departamento de Ingenier´ıaEl´ectrica,Electr´onicay Computaci´on Grupo de Procesamiento Digital de Senales Enero 2014 UNIVERSIDAD NACIONAL DE COLOMBIA Abstract Faculty of Engineering and Architecture Department of Electronic, Electric Engineering and Computation Master’s on Engineering - Industrial Automation Methodology for predicting semantic annotations of protein sequences by feature extraction derived of statistical contact potentials and continuous wavelet transform by Gustavo Alonso Arango Argoty In this thesis, a method to predict semantic annotations of the proteins from its primary structure is proposed. The main contribution of this thesis lies in the implementation of a novel protein feature representation, which makes use of the pairwise statistical contact potentials describing the protein interactions and geometry at the atomic level. -

Genbank by Walter B
GenBank by Walter B. Goad o understand the significance of the information stored in memory—DNA—to the stuff of activity—proteins. The idea that GenBank, you need to know a little about molecular somehow the bases in DNA determine the amino acids in proteins genetics. What that field deals with is self-replication—the had been around for some time. In fact, George Gamow suggested in T process unique to life—and mutation and 1954, after learning about the structure proposed for DNA, that a recombination—the processes responsible for evolution-at the triplet of bases corresponded to an amino acid. That suggestion was fundamental level of the genes in DNA. This approach of working shown to be true, and by 1965 most of the genetic code had been from the blueprint, so to speak, of a living system is very powerful, deciphered. Also worked out in the ’60s were many details of what and studies of many other aspects of life—the process of learning, for Crick called the central dogma of molecular genetics—the now example—are now utilizing molecular genetics. firmly established fact that DNA is not translated directly to proteins Molecular genetics began in the early ’40s and was at first but is first transcribed to messenger RNA. This molecule, a nucleic controversial because many of the people involved had been trained acid like DNA, then serves as the template for protein synthesis. in the physical sciences rather than the biological sciences, and yet These great advances prompted a very distinguished molecular they were answering questions that biologists had been asking for geneticist to predict, in 1969, that biology was just about to end since years. -

Deep Learning in Chemoinformatics Using Tensor Flow
UC Irvine UC Irvine Electronic Theses and Dissertations Title Deep Learning in Chemoinformatics using Tensor Flow Permalink https://escholarship.org/uc/item/963505w5 Author Jain, Akshay Publication Date 2017 Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California UNIVERSITY OF CALIFORNIA, IRVINE Deep Learning in Chemoinformatics using Tensor Flow THESIS submitted in partial satisfaction of the requirements for the degree of MASTER OF SCIENCE in Computer Science by Akshay Jain Thesis Committee: Professor Pierre Baldi, Chair Professor Cristina Videira Lopes Professor Eric Mjolsness 2017 c 2017 Akshay Jain DEDICATION To my family and friends. ii TABLE OF CONTENTS Page LIST OF FIGURES v LIST OF TABLES vi ACKNOWLEDGMENTS vii ABSTRACT OF THE THESIS viii 1 Introduction 1 1.1 QSAR Prediction Methods . .2 1.2 Deep Learning . .4 2 Artificial Neural Networks(ANN) 5 2.1 Artificial Neuron . .5 2.2 Activation Function . .7 2.3 Loss function . .8 2.4 Optimization . .8 3 Deep Recursive Architectures 10 3.1 Recurrent Neural Networks (RNN) . 10 3.2 Recursive Neural Networks . 11 3.3 Directed Acyclic Graph Recursive Neural Networks (DAG-RNN) . 11 4 UG-RNN for small molecules 14 4.1 DAG Generation . 16 4.2 Local Information Vector . 16 4.3 Contextual Vectors . 17 4.4 Activity Prediction . 17 4.5 UG-RNN With Contracted Rings (UG-RNN-CR) . 18 4.6 Example: UG-RNN Model of Propionic Acid . 20 5 Implementation 24 6 Data & Results 26 6.1 Aqueous Solubility Prediction . 26 6.2 Melting Point Prediction . 28 iii 7 Conclusions 30 Bibliography 32 A Source Code 37 A.1 UGRNN . -

Course Outline
Department of Computer Science and Software Engineering COMP 6811 Bioinformatics Algorithms (Reading Course) Fall 2019 Section AA Instructor: Gregory Butler Curriculum Description COMP 6811 Bioinformatics Algorithms (4 credits) The principal objectives of the course are to cover the major algorithms used in bioinformatics; sequence alignment, multiple sequence alignment, phylogeny; classi- fying patterns in sequences; secondary structure prediction; 3D structure prediction; analysis of gene expression data. This includes dynamic programming, machine learning, simulated annealing, and clustering algorithms. Algorithmic principles will be emphasized. A project is required. Outline of Topics The course will focus on algorithms for protein sequence analysis. It will not cover genome assembly, genome mapping, or gene recognition. • Background in Biology and Genomics • Sequence Alignment: Pairwise and Multiple • Representation of Protein Amino Acid Composition • Profile Hidden Markov Models • Specificity Determining Sites • Curation, Annotation, and Ontologies • Machine Learning: Secondary Structure, Signals, Subcellular Location • Protein Families, Phylogenomics, and Orthologous Groups • Profile-Based Alignments • Algorithms Based on k-mers 1 Texts | in Library D. Higgins and W. Taylor (editors). Bioinformatics: Sequence, Structure and Databanks, Oxford University Press, 2000. A. D. Baxevanis and B. F. F. Ouelette. Bioinformatics: A Practical Guide to the Analysis of Genes and Proteins, Wiley, 1998. Richard Durbin, Sean R. Eddy, Anders Krogh, -

Integrative Annotation of 21,037 Human Genes Validated by Full-Length Cdna Clones
Title Integrative Annotation of 21,037 Human Genes Validated by Full-Length cDNA Clones Imanishi, Tadashi; Itoh, Takeshi; Suzuki, Yutaka; O'Donovan, Claire; Fukuchi, Satoshi; Koyanagi, Kanako O.; Barrero, Roberto A.; Tamura, Takuro; Yamaguchi-Kabata, Yumi; Tanino, Motohiko; Yura, Kei; Miyazaki, Satoru; Ikeo, Kazuho; Homma, Keiichi; Kasprzyk, Arek; Nishikawa, Tetsuo; Hirakawa, Mika; Thierry-Mieg, Jean; Thierry-Mieg, Danielle; Ashurst, Jennifer; Jia, Libin; Nakao, Mitsuteru; Thomas, Michael A.; Mulder, Nicola; Karavidopoulou, Youla; Jin, Lihua; Kim, Sangsoo; Yasuda, Tomohiro; Lenhard, Boris; Eveno, Eric; Suzuki, Yoshiyuki; Yamasaki, Chisato; Takeda, Jun-ichi; Gough, Craig; Hilton, Phillip; Fujii, Yasuyuki; Sakai, Hiroaki; Tanaka, Susumu; Amid, Clara; Bellgard, Matthew; Bonaldo, Maria de Fatima; Bono, Hidemasa; Bromberg, Susan K.; Brookes, Anthony J.; Bruford, Elspeth; Carninci, Piero; Chelala, Claude; Couillault, Christine; Souza, Sandro J. de; Debily, Marie-Anne; Devignes, Marie-Dominique; Dubchak, Inna; Endo, Toshinori; Estreicher, Anne; Eyras, Eduardo; Fukami-Kobayashi, Kaoru; R. Gopinath, Gopal; Graudens, Esther; Hahn, Yoonsoo; Han, Michael; Han, Ze-Guang; Hanada, Kousuke; Hanaoka, Hideki; Harada, Erimi; Hashimoto, Katsuyuki; Hinz, Ursula; Hirai, Momoki; Hishiki, Teruyoshi; Hopkinson, Ian; Imbeaud, Sandrine; Inoko, Hidetoshi; Kanapin, Alexander; Kaneko, Yayoi; Kasukawa, Takeya; Kelso, Janet; Kersey, Author(s) Paul; Kikuno, Reiko; Kimura, Kouichi; Korn, Bernhard; Kuryshev, Vladimir; Makalowska, Izabela; Makino, Takashi; Mano, Shuhei; -

Deep Learning in Science Pierre Baldi Frontmatter More Information
Cambridge University Press 978-1-108-84535-9 — Deep Learning in Science Pierre Baldi Frontmatter More Information Deep Learning in Science This is the first rigorous, self-contained treatment of the theory of deep learning. Starting with the foundations of the theory and building it up, this is essential reading for any scientists, instructors, and students interested in artificial intelligence and deep learning. It provides guidance on how to think about scientific questions, and leads readers through the history of the field and its fundamental connections to neuro- science. The author discusses many applications to beautiful problems in the natural sciences, in physics, chemistry, and biomedicine. Examples include the search for exotic particles and dark matter in experimental physics, the prediction of molecular properties and reaction outcomes in chemistry, and the prediction of protein structures and the diagnostic analysis of biomedical images in the natural sciences. The text is accompanied by a full set of exercises at different difficulty levels and encourages out-of-the-box thinking. Pierre Baldi is Distinguished Professor of Computer Science at University of California, Irvine. His main research interest is understanding intelligence in brains and machines. He has made seminal contributions to the theory of deep learning and its applications to the natural sciences, and has written four other books. © in this web service Cambridge University Press www.cambridge.org Cambridge University Press 978-1-108-84535-9 — Deep Learning in -

Bringing Folding Pathways Into Strand Pairing Prediction
Bringing folding pathways into strand pairing prediction Jieun Jeong1,2, Piotr Berman1, and Teresa Przytycka2 1 Computer Science and Engineering Department The Pennsylvania State University University Park, PA 16802 USA 2 National Center for Biotechnology Information US National Library of Medicine, National Institutes of Health Bethesda, MD 20894 email: [email protected], [email protected], [email protected] Abstract. The topology of β-sheets is defined by the pattern of hydrogen- bonded strand pairing. Therefore, predicting hydrogen bonded strand partners is a fundamental step towards predicting β-sheet topology. In this work we report a new strand pairing algorithm. Our algorithm at- tempts to mimic elements of the folding process. Namely, in addition to ensuring that the predicted hydrogen bonded strand pairs satisfy basic global consistency constraints, it takes into account hypothetical folding pathways. Consistently with this view, introducing hydrogen bonds be- tween a pair of strands changes the probabilities of forming other strand pairs. We demonstrate that this approach provides an improvement over previously proposed algorithms. 1 Introduction The prediction of protein structure from protein sequence is a long-held goal that would provide invaluable information regarding the function of individ- ual proteins and the evolution of protein families. The increasing amount of sequence and structure data, made it possible to decouple the structure predic- tion problem from the problem of modeling of protein folding process. Indeed, a significant progress has been achieved by bioinformatics approaches such as homology modeling, threading, and assembly from fragments [16]. At the same time, the fundamental problem of how actually a protein acquires its final folded state remains a subject of controversy. -
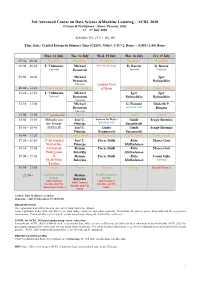
ACDL-2020-Programme-Ver10.0
3rd Advanced Course on Data Science &Machine Learning – ACDL 2020 Certosa di Pontignano - Siena, Tuscany, Italy 13 – 17 July 2020 Schedule Ver. 11.0 – July 9th Time Zone: Central European Summer Time (CEST), Offset: UTC+2, Rome – (GMT+2:00) Rome Mon, 13 July Tue, 14 July Wed, 15 July Thu, 16 July Fri, 17 July 07:30 – 09:00 Breakfast Breakfast Breakfast Breakfast Breakfast 09:00 – 09:50 T. Viehmann Michael 8:30 Social Tour D. Bacciu D. Bacciu Tutorial Bronstein Tutorial Tutorial 09:50 – 10:40 Michael Igor Bronstein Babuschkin Tutorial Guided Visit 10:40 – 11:20 Coffee break Coffee break of Siena Coffee break Coffee break 11:20 – 12:10 T. Viehmann Michael Igor Igor Tutorial Bronstein Babuschkin Babuschkin Tutorial 12:10 – 13:00 Michael G. Fiameni Diederik P. Bronstein Industrial Talk Kingma Tutorial 13:00 – 15:00 Lunch (13:00-14:00) Lunch Lunch Lunch Lunch 15:00 – 15:50 Mihaela van José C. Lorenzo De Mattei Guido Sergiy Butenko der Schaar Principe Industrial Talk Sanguinetti 15:50 – 16:40 (14:00-16:40) José C. Guido Guido Sergiy Butenko Principe Sanguinetti Sanguinetti 16:40 – 17:20 Coffee break Coffee break Coffee break Coffee break Coffee break 17:20 – 18:10 17:20 Guided José C. Pierre Baldi Risto Marco Gori Visit of the Principe Miikkulainen 18:10 – 19:00 Certosa di Roman Pierre Baldi Risto Marco Gori Pontignano Belavkin Miikkulainen 19:00 – 19:50 & Roman Pierre Baldi Risto Varun Ojha 18:20 Wine Belavkin Miikkulainen Tutorial Tasting 19:50 – 21:50 Dinner Dinner Dinner Dinner Social Dinner 21:50 – Oral Presentation Roman Oral Presentation Session Belavkin Session with Cantucci with Cantucci with Cantucci biscuits and Vin biscuits and Vin biscuits and Vin Santo (sweet wine) Santo Santo Arrival: July 12 (Dinner at 20:30) Departure: July 18 (Breakfast 07:30-09:00) REGISTRATION The registration desk will be located close to the Main Conference Room.