Modeling Spatiotemporal Patterns of PM2.5 at the Sub-Neighborhood Scale Using Low-Cost Sensor Networks
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Air Quality Monitoring Toolkit: Assessing Second-Hand Smoke in Hospitality Venues
Air Quality Monitoring Toolkit: Assessing Second-Hand Smoke in Hospitality Venues Authors: Dr Angela Jackson-Morris, Department of Tobacco Control, The International Union Against Tuberculosis and Lung Disease, Edinburgh, Scotland; Dr Sean Semple, Scottish Centre for Indoor Air, Respiratory Group, Division of Applied Health Sciences, University of Aberdeen, Aberdeen, Scotland; Ruaraidh Dobson, Scottish Centre for Indoor Air, Child Health, University of Aberdeen, Aberdeen, Scotland. About the International Union Against Tuberculosis and Lung Disease (The Union): For nearly 100 years, The Union has drawn from the best scientific evidence and the skills, expertise and reach of its staff, consultants and membership in order to advance solutions to the most pressing public health challenges affecting people living in poverty around the world. With nearly 17,000 members and subscribers from 156 countries, The Union has its headquarters in Paris and regional offices in Africa, the Asia Pacific, Europe, Latin America, North America and South-East Asia. The Union’s scientific departments focus on tuberculosis and HIV, lung health and non- communicable diseases, tobacco control and operational research. For more information on The Union’s tobacco control work please visit www.tobaccofreeunion.org or follow us on Twitter @TheUnion_TC. Contact: The International Union Against Tuberculosis and Lung Disease (The Union) Department of Tobacco Control 8 Randolph Crescent Edinburgh UK EH3 7TH T: 0131 240 0252 E: [email protected] About the University of Aberdeen: Founded in 1495, the University of Aberdeen is Scotland's third oldest and the UK's fifth oldest university, and is consistently ranked among the top 1 percent of the world's universities. -

Open Source on IBM I Webinar Series Day 2 ERWIN EARLEY ([email protected]), SR
Open Source on IBM i Webinar Series Day 2 ERWIN EARLEY ([email protected]), SR. SOLUTIONS CONSULTANT, PERFORCE, NOVEMBER 2019 2 | COMMON Webinar Series: Open Source on IBM i | November 2019 zend.com Day 1 Review • Introduction to Open Source on IBM i • Why is Open Source on IBM i Important • Understanding the PASE environment as the enabler of Open Source on IBM i • Getting Familiar with the PASE environment 2 | Zend by Perforce © 2019 Perforce Software, Inc. zend.com 3 | COMMON Webinar Series: Open Source on IBM i | November 2019 zend.com Day 2 Agenda • Setting up OSS EcoSystem on IBM i – ACS version • Exploring Containers on IBM i • Managing Open Source on IBM i • Exploring Open Source Programming Languages ▪ Integration with Db2 and ILE • After-Hours Lab: Containers & Setting up Development Environment • After-Hours Lab: Open Source Programming Languages 3 | Zend by Perforce © 2019 Perforce Software, Inc. zend.com IBM Systems Technical University © 3 4 | COMMON Webinar Series: Open Source on IBM i | November 2019 zend.com Setting up OSS Ecosystem on IBM i – ACS Version 4 | Zend by Perforce © 2019 Perforce Software, Inc. zend.com 5 | COMMON Webinar Series: Open Source on IBM i | November 2019 zend.com The directory structure Before installing the Open Source ecosystem / dev home lib sbin tmp usr var Directory Contents bin Commands dev Device Files etc Configuration files home User Home Directories lib Libraries pkgs Package files / commands sbin Privileged commands tmp Temporary files usr Utilities & Applications var Variable files -

Server-Side Rendering of React Applications in Enterprise Portals
Bachelor’s Thesis Czech Technical University in Prague Faculty of Electrical Engineering F3 Department of Computer Science Server-side rendering of React applications in enterprise portals Václav Jančařík Supervisor: Ing. Martin Ledvinka Field of study: Software Engineering and Technology May 2019 ii ZADÁNÍ BAKALÁŘSKÉ PRÁCE I. OSOBNÍ A STUDIJNÍ ÚDAJE Příjmení: Jančařík Jméno: Václav Osobní číslo: 466301 Fakulta/ústav: Fakulta elektrotechnická Zadávající katedra/ústav: Katedra počítačů Studijní program: Softwarové inženýrství a technologie II. ÚDAJE K BAKALÁŘSKÉ PRÁCI Název bakalářské práce: Vykreslování React aplikací na straně serveru v enterprise portálech Název bakalářské práce anglicky: Server-side rendering of React applications in enterprise portals Pokyny pro vypracování: 1. Analyze the current state of the art in the field of server-side rendering of React applications and running React applications in portal solutions. 2. Design fundamental principles of integration of server-side rendering of React applications in the context of portal environments. 3. Based on your design, implement a server-side rendering solution for React applications embedded in enterprise portals. 4. Demonstrate the correctness of your solution by comparing client- side and server-side rendering output of an example application. 5. Compare the performance of your server-side rendering solution with standard client-side rendering. Seznam doporučené literatury: [1] K. Konshin, Next.js Quick Start Guide: Server-side rendering done right, Packt Publishing, 2018 [2] R. Sezov, Liferay in Action: The Official Guide to Liferay Portal Development, Manning Publications, 2011 [3] R. Wieruch, The Road to learn React: Your journey to master plain yet pragmatic React.js, 2018 Jméno a pracoviště vedoucí(ho) bakalářské práce: Ing. -

Adapt Authoring Tool: Installation
Adapt Authoring Tool: Installation UGA Training Capstone Team April 21, 2019 1 Contents 1 Installing Prerequisites 3 1.1 Installing Git . 3 1.1.1 Debian Based Install . 3 1.1.2 RPM Based Install . 3 1.2 NodeJS . 4 1.2.1 Debian Based Install . 4 1.2.2 RPM Based Install . 4 1.3 Grunt . 5 1.3.1 Update NPM . 5 1.3.2 Install Grunt . 5 1.4 MongoDB Community Edition . 6 1.4.1 Debian Based Install . 6 1.4.2 RPM Based Install . 6 1.5 FFmpeg . 7 1.5.1 Debian Based Install . 7 1.5.2 RPM Based Install . 7 2 Installing The Authoring Tool 8 2.1 Clone Adapt Authoring . 8 2.2 Install required NPM packages . 8 2.3 Install Script . 9 2.4 PM2: Process Manager . 11 2.4.1 Installing PM2 . 11 2.4.2 Starting The Server . 11 2 1 Installing Prerequisites The authoring tool and framework require other software to operate. This section will provide the directions on what to install. Instructions have been provided for installation on a Debian or RPM based server. Other instruc- tions are provided at each software packages website. Administrative or Sudo access is required to complete the installation. 1.1 Installing Git Git is a tool for managing source code, and makes it easier to download and update the software. Git may already be installed on the server to which you are installing the authoring tool, to check from command line: $ git --version. 1.1.1 Debian Based Install From the command line: $ sudo apt install git-all 1.1.2 RPM Based Install From the command line: $ sudo dnf install git-all 3 1.2 NodeJS NodeJS is an open source, cross-platform JavaScript run-time enviroment that executes JavaScript code outside of a browser. -
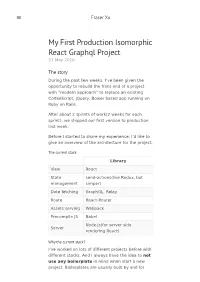
My First Production Isomorphic React Graphql Project 31 May 2016
Fraser Xu My First Production Isomorphic React Graphql Project 31 May 2016 The story During the past few weeks, I’ve been given the opportunity to rebuild the front-end of a project with “modern approach” to replace an existing CoffeeScript, jQuery, Bower based app running on Ruby on Rails. After about 2 sprints of work(2 weeks for each sprint), we shipped our first version to production last week. Before I started to share my experience, I’d like to give an overview of the architecture for the project. The current stack Library View React State send-actions(like Redux, but management simper) Date fetching GraphQL, Relay Route React-Router Assets serving Webpack Precompile JS Babel Node.js(for server side Server rendering React) Why the current stack? I’ve worked on lots of different projects before with different stacks. And I always have the idea to not use any boilerplate in mind when start a new project. Boilerplates are usually built by and for people with different requirements for a project, and none of them are identical to the one you are trying to build. So usually I will only keep a list of well maintained boilerplate project, and only use them as a reference when my own stack gets into trouble. The new project has a few requirements: Server side rendering for progressive enhanced experience so the page could work for user without JavaScript SEO, we are mainly an e-commercial website, so SEO is the number one priority The app needs to talk to a couple of micro- services, and tokens are usually stored on the server for safety reasons UI state should persist from url, not only for SEO, but also for a better user experience Fast iteration time, to move fast and delivery better user experience Improve performance, the short time we delivery page to user, the longer we can keep the user on the website There are also other requirements which are not for business, and most of them are actually for a better developer experience. -

Portfolio Appendix to the Full CV
Portfolio Appendix to the full CV Tobias Cudnik Fullstack Web Developer [email protected] github.com/TobiaszCudnik linkedin.com/in/tobiaszcudnik Table of Contents 1. Node.js (TS / JS) 2. JavaScript (FE) 3. TypeScript (FE) 4. PHP 5. Golang 6. Python 7. Mobile 8. Automated testing 9. DevOps 10. Product design 11. Remote work 12. Showcase projects Latest version at https://bendedlogic.com/portfolio 1. Node.js (TS / JS) ● Closed Source ○ UBIO TypeScript, Koa, MongoDB, Microservices, Web Scraping, Google Travel API ○ Hola Express, MongoDB, p2p, LXC, CVS, ES3->ES6, custom tools, ESLint plugins ○ BetVictor Express, MongoDB, SQL, Microservices ● Commercial Open Source ○ https://github.com/RiseVision/rise-node (commits) TypeScript, Blockchain, CLI, P2P, Docker, Protocol Buffers, nginx, Distributed System ● Open Source ○ https://github.com/TobiaszCudnik/taskbot TypeScript, Next.js, Hapi.js, Google Tasks API, GMail API, Google Sign-In, Firebase, AsyncMachine ○ https://github.com/TobiaszCudnik/asyncmachine-inspector TypeScript, Workers, RPC, JSONDiffPatch, Redis, Socket.io ○ https://github.com/TobiaszCudnik/wsti-thesis-2012 P2P, RPC, Distributed System, Graph Theory 2. JavaScript (FE) ● Closed Source ○ Hola Angular, MongoDB, ES6, CVS ○ William Hill React, Redux, Redux Sagas, Flowtype, REST, Git ○ CloudFarm5 Backbone, Chaplin, REST, data visualization (Canvas), CoffeeScript, ES5, jQuery, Git ○ Google Closure Compiler, Closure Library, Protocol Buffers, JSDoc, Vanilla JS, ES3, Perforce, Git ○ UBS UI optimization, refactoring, Backbone, Mustache, -

Complete Node.Js Secrets & Tips for Professionals
Node.js CompleteComplete Tips & Secrets for Professionals Node.js Tips & Secrets for Professionals 200+ pages of professional hints and tricks Disclaimer This is an unocial free book created for educational purposes and is GoalKicker.com not aliated with ocial Node.js group(s) or company(s). Free Programming Books All trademarks and registered trademarks are the property of their respective owners Contents About ................................................................................................................................................................................... 1 Chapter 1: Getting started with Node.js ............................................................................................................ 2 Section 1.1: Hello World HTTP server ........................................................................................................................... 3 Section 1.2: Hello World command line ....................................................................................................................... 4 Section 1.3: Hello World with Express .......................................................................................................................... 5 Section 1.4: Installing and Running Node.js ................................................................................................................. 6 Section 1.5: Debugging Your NodeJS Application ...................................................................................................... 6 Section 1.6: -

Pavel Polyakov's Blog
Pavel Polyakov Germany, Hamburg, 22087 Immenhof 11 +4915258163147 [email protected] skype: pavel.polyakov.x1 Personal blog: http://pavelpolyakov.com Linkedin: https://www.linkedin.com/in/pavel-polyakov Github: https://github.com/PavelPolyakov Education Kharkiv State Economics University Master’s degree, Computer Science 2004 – 2009 Grade: A Work Experience Kreditech, April 2015 - present ● Expert Software Engineer, developing using Node.js, mainly Hapi.js framework, Angular 1 as front-end. React as of 2017. Responsible as for the architecture (partly) as for bringing the project live. * Appendix A X1 Group, August 2012 – April 2015 ● Senior PHP developer, developing Zend Framework 1/Laravel 4 based applications from the scratch, developing the project architecture, integrating various API. * Appendix A Total Internet Group, August 2009 – August 2012 ● Senior PHP developer / Office leader, developing Zend Framework based projects and Magento based web shops. Leading small teams in the way to make the projects successful. Also I was responsible for the office functioning in general. -

Jiskefet, a Bookkeeping Application for ALICE
EPJ Web of Conferences 245, 04023 (2020) https://doi.org/10.1051/epjconf/202024504023 CHEP 2019 Jiskefet, a bookkeeping application for ALICE 1,* 2 1 Marten Teitsma , Vasco Chibante Barosso , Pascal Boeschoten , and Patrick Hendriks1 1Amsterdam University of Applied Sciences 2CERN Abstract. A new bookkeeping system called Jiskefet is being developed for A Large Ion Collider Experiment (ALICE) during Long Shutdown 2, to be in production until the end of LHC Run 4 (2029). Jiskefet unifies two function- alities: a) gathering, storing and presenting metadata associated with the oper- ations of the ALICE experiment and b) tracking the asynchronous processing of the physics data. It will replace the existing ALICE Electronic Logbook and AliMonitor, allowing for a technology refresh and the inclusion of new fea- tures based on the experience collected during Run 1 and Run 2. The front end leverages web technologies much in use nowadays such as TypeScript and NodeJS and is adaptive to various clients such as tablets, mobile devices and other screens. The back end includes an OpenAPI specification based REST API and a relational database. This paper will describe the organization of the work done by various student teams who work on Jiskefet in sequential and parallel semesters and how continuity is guaranteed by using guidelines on cod- ing, documentation and development. It will also describe the current status of the development, the initial experience in detector stand-alone commissioning setups and the future plans. 1 Introduction During the LHC Long Shutdown 2 a renewal of the bookkeeping systems in place for AL- ICE is envisioned. -

경력사항 국방통합데이터센터 1센터 - 만기제대 2014-08-11 ~ 2016-05-10 - 군대�Sw 관리병� / 체계관제병 - 서버 모니터링 및 유지보수� 스크립트 실행하여 서버 재실행 등
함께하면 서로 성장하는 개발자 정석호입니다. 정석호 httpsvuugithubtcomuCreatiCoding / nodejsdeveloper@kakaotcom 자기소개 안녕하세요r 저는 Viva Republica에서 Client Platform에서 일하고 있는 Frontend DeveloperpDevopsq 입니다t frontsend 개발자도 DevOps를 조금만 알면 backsend 개발자 못지않게 유용하게 활용할 수 있음을 널리 알리고 싶습니다t #DivOps는 재밌어요 ✨ 저는 아래를 추구하고l고민합니다t - 최소한의 리소스로 정직하고 최대한의 효과 - 성능 좋은 코드 < 읽기 좋은 코드 - 주석 달린 코드 < 직관적인 코드 - 개발자 모두의e상향 평준화 저는 이것을 좋아합니다t - frontsend를 위한eDevOpsp자동배포 및 유닛테스트q - 성능 최적화p첫 로딩 속도r 번들링 분석q - 트러블 슈팅p레이아웃 쉬프트r 플리커l현상) 경력사항 국방통합데이터센터 1센터 - 만기제대 2014-08-11 ~ 2016-05-10 - 군대pSW 관리병q / 체계관제병 - 서버 모니터링 및 유지보수r 스크립트 실행하여 서버 재실행 등 웍스모바일 - 인턴 8주 2018-07-02 ~ 2018-08-24 - 채용전제형 인턴 / 인턴 과제 진행 - Elasticsearch를 활용한 검색엔진 개선 pElasticsearchr Logstashr c언어q 펫프렌즈 - 퇴사 2018-12-17 ~ 2021-05-21 - 정규직 / Frontend Developer - 신규 서비스 UI 담당 및 기존 서비스 유지보수 비바리퍼블리카 - 재직 중 2021-05-31 ~ - 정규직 / Frontend Developer pdevopsq - 프론트엔드 개발 환경 개선 및 CIuCD 안정화 경력 기술서 feature 긴급점검 기능 2021-03 ~ 2021-03 - Nodetjs Koa의 middleware 기능으로 redirect response 추가 - axios의 interceptor에서 301을 찾아 response의 redirect URL 로 변경 - Backend의 response의 데이터 중 URL에 query string 추가하여 점검 시간을 동적 데이터로 표 기 - 기존 방식v 이미지 교체 + 정적 소스 배포 - => 디자이너 + 프론트엔드 개발자의 리소스를 절약 edu 사내 교육 frontsend 신입 개발자 Vuetjs 세미나 2021-03 ~ - frontsend 신입 개발자를 위한 사내 개발팀 세미나를 진행 중 - 공통 코드 스타일 정립 및 공유 - httpsvuugithubtcomuCreatiCodinguseminarsvuejs project 신 파트너사 사이트 개발 pReact Nexttjs 무중단 배포q new 2020-08 ~ 2020-11 - 파트너사를 위한 웹 사이트를 React Nexttjs 기반의 SSR로 진행 - 2대의 서버에 각 서버에서 pm2 를 활용하여 무중단 배포 - 상용환경은 서버 2대에 로드밸런싱을 적용했으며r 그로 인해 발생했던 사이드이펙트도 해결 feature 배너 움짤 2020-06 ~ 2020-07 - 기존 -

Brenda Zhang
[email protected] Brenda Zhang 415-312-3215 Cog Sci Major & Comp Sci Minor linkedin.com/in/brendacs University of California, Berkeley github.com/brendacs 08/2015 - 05/2019 (anticipated) brendacs.github.io/desktop WORK EXPERIENCE TECHNICAL SKILLS Software Engineering Intern Languages & Databases HTML5, CSS3, JavaScript, Python, Java, SQL, SOQL, The Climate Corporation Apex, PostgreSQL, MongoDB, Salesforce 05/2018 - 08/2018 San Francisco, CA Frameworks & Libraries • Developed React Native app, for internal staff to scan QR React, React Native, Angular, Redux, Node.js, Ex- codes, which queries/updates Salesforce records using SQL press, Rails, Django, jQuery, Next.js • Modernized events page by creating search, sort, and edit Miscellaneous Tools, Managers & Preprocessors Java functions and added form with Java/SQL backend SASS, LESS, Grunt, Gulp, webpack, npm, Yarn, pip, • Enhanced testing automation with addition of over 100 front Postman, Git, GitHub, PM2, DigitalOcean, Heroku and back -end unit tests written in Java, Jest, and Enzyme • Added reusable React feature components, such as toast no- Testing Frameworks & Tools tifications, tooltips, and components for internationalization Jest, Enzyme, TestCafe, unittest, JUnit Full Stack Engineer PERSONAL PROJECTS Stowk, Inc. 06/2017 - Present 03/2017 - 05/2018 Berkeley, CA Discord Stop Bot • A message moderation bot for Discord that automatically stops • Built and maintained the front end of the web and cross-plat- spam, profanity, and other unwanted messages, especially in large servers. It was developed with Node.js, Express, Post- form mobile apps using React, React Native, and Redux greSQL, and Discord.js. Managed with PM2 on VPS. It is active • Developed shipment progress dashboard, facial recognition in over 2,400 Discord servers with over 95,000 users. -

Health Risks of Particulate Matter from Long-Range Transboundary Pollution
Particulate matter is a type of air pollution that The WHO Regional Office transboundary airpollution long-range from Health risksofparticulate matter for Europe is generated by a variety of human activities, transboundary airpollution long-range Health risksofparticulate from matter can travel long distances in the atmosphere and The World Health Organization (WHO) is a specialized agency of the United causes a wide range of diseases and a significant Nations created in 1948 with the primary reduction of life expectancy in most of the responsibility for international health matters and public health. The WHO population of Europe. Regional Office for Europe is one of six regional offices throughout the world, each with its own programme geared to This report summarizes the evidence on these the particular health conditions of the countries it serves. effects, as well as knowledge about the sources of particulate matter, its transport in the Member States atmosphere, measured and modelled levels Albania Andorra of pollution in ambient air, and population Armenia Austria exposure. It shows that long-range transport of Azerbaijan particulate matter contributes significantly to Belarus Belgium exposure and to health effects. Bosnia and Herzegovina Bulgaria Croatia The authors conclude that international action Cyprus Czech Republic must accompany local and national efforts to cut Denmark pollution emissions and reduce their effects on Estonia Finland human health. France Georgia Germany Greece Hungary Iceland Ireland Israel Italy Kazakhstan Kyrgyzstan Latvia Lithuania Luxembourg Malta Monaco Netherlands Norway Poland Portugal Republic of Moldova Romania Russian Federation San Marino Serbia and Montenegro Slovakia Slovenia Spain Sweden World Health Organization Switzerland Regional Office for Europe Tajikistan The former Yugoslav Republic of Macedonia Scherfigsvej 8, DK-2100 Copenhagen Ø, Denmark Turkey Tel.: +45 39 17 17 17.