Development and Characterization of Fungal Specific Microsatellite
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Macroevolutionary Dynamics of Symbiotic and Phenotypic Diversification in Lichens
The macroevolutionary dynamics of symbiotic and phenotypic diversification in lichens Matthew P. Nelsena,1, Robert Lückingb, C. Kevin Boycec, H. Thorsten Lumbscha, and Richard H. Reea aDepartment of Science and Education, Negaunee Integrative Research Center, The Field Museum, Chicago, IL 60605; bBotanischer Garten und Botanisches Museum, Freie Universität Berlin, 14195 Berlin, Germany; and cDepartment of Geological Sciences, Stanford University, Stanford, CA 94305 Edited by Joan E. Strassmann, Washington University in St. Louis, St. Louis, MO, and approved July 14, 2020 (received for review February 6, 2020) Symbioses are evolutionarily pervasive and play fundamental roles macroevolutionary consequences of ant–plant interactions (15–19). in structuring ecosystems, yet our understanding of their macroevo- However, insufficient attention has been paid to one of the most lutionary origins, persistence, and consequences is incomplete. We iconic examples of symbiosis (20, 21): Lichens. traced the macroevolutionary history of symbiotic and phenotypic Lichens are stable associations between a mycobiont (fungus) diversification in an iconic symbiosis, lichens. By inferring the most and photobiont (eukaryotic alga or cyanobacterium). The pho- comprehensive time-scaled phylogeny of lichen-forming fungi (LFF) tobiont supplies the heterotrophic fungus with photosynthetically to date (over 3,300 species), we identified shifts among symbiont derived carbohydrates, while the mycobiont provides the pho- classes that broadly coincided with the convergent -

Lichens of East Limestone Island
Lichens of East Limestone Island Stu Crawford, May 2012 Platismatia Crumpled, messy-looking foliose lichens. This is a small genus, but the Pacific Northwest is a center of diversity for this genus. Out of the six species of Platismatia in North America, five are from the Pacific Northwest, and four are found in Haida Gwaii, all of which are on Limestone Island. Platismatia glauca (Ragbag lichen) This is the most common species of Platismatia, and is the only species that is widespread. In many areas, it is the most abundant lichen. Oddly, it is not the most abundant Platismatia on Limestone Island. It has soredia or isidia along the edges of its lobes, but not on the upper surface like P. norvegica. It also isn’t wrinkled like P. norvegica or P. lacunose. Platismatia norvegica It has large ridges or wrinkles on its surface. These ridges are covered in soredia or isidia, particularly close to the edges of the lobes. In the interior, it is restricted to old growth forests. It is less fussy in coastal rainforests, and really seems to like Limestone Island, where it is the most abundant Platismatia. Platismatia lacunosa (wrinkled rag lichen) This species also has large wrinkles on its surface, like P. norvegica. However, it doesn’t have soredia or isidia on top of these ridges. Instead, it has tiny black dots along the edges of its lobes which produce spores. It is also usually whiter than P. lacunosa. It is less common on Limestone Island. Platismatia herrei (tattered rag lichen) This species looks like P. -

Extended Phylogeny and a Revised Generic Classification of The
The Lichenologist 46(5): 627–656 (2014) 6 British Lichen Society, 2014 doi:10.1017/S002428291400019X Extended phylogeny and a revised generic classification of the Pannariaceae (Peltigerales, Ascomycota) Stefan EKMAN, Mats WEDIN, Louise LINDBLOM and Per M. JØRGENSEN Abstract: We estimated phylogeny in the lichen-forming ascomycete family Pannariaceae. We specif- ically modelled spatial (across-site) heterogeneity in nucleotide frequencies, as models not incorpo- rating this heterogeneity were found to be inadequate for our data. Model adequacy was measured here as the ability of the model to reconstruct nucleotide diversity per site in the original sequence data. A potential non-orthologue in the internal transcribed spacer region (ITS) of Degelia plumbea was observed. We propose a revised generic classification for the Pannariaceae, accepting 30 genera, based on our phylogeny, previously published phylogenies, as well as available morphological and chemical data. Four genera are established as new: Austroparmeliella (for the ‘Parmeliella’ lacerata group), Nebularia (for the ‘Parmeliella’ incrassata group), Nevesia (for ‘Fuscopannaria’ sampaiana), and Pectenia (for the ‘Degelia’ plumbea group). Two genera are reduced to synonymy, Moelleropsis (included in Fuscopannaria) and Santessoniella (non-monophyletic; type included in Psoroma). Lepido- collema, described as monotypic, is expanded to include 23 species, most of which have been treated in the ‘Parmeliella’ mariana group. Homothecium and Leightoniella, previously treated in the Collemataceae, are here referred to the Pannariaceae. We propose 41 new species-level combinations in the newly described and re-circumscribed genera mentioned above, as well as in Leciophysma and Psoroma. Key words: Collemataceae, lichen taxonomy, model adequacy, model selection Accepted for publication 13 March 2014 Introduction which include c. -

Taxonomy, Phylogeny and Biogeography of the Lichen Genus Peltigera in Papua New Guinea
Fungal Diversity Taxonomy, phylogeny and biogeography of the lichen genus Peltigera in Papua New Guinea Sérusiaux, E.1*, Goffinet, B.2, Miadlikowska, J.3 and Vitikainen, O.4 1Plant Taxonomy and Conservation Biology Unit, University of Liège, Sart Tilman B22, B-4000 Liège, Belgium 2Department of Ecology and Evolutionary Biology, University of Connecticut, 75 North Eagleville Road, Storrs CT 06269-3043 USA 3Department of Biology, Duke University, Durham, NC 27708-0338, USA 4Botanical Museum (Mycology), P.O. Box 7, FI-00014 University of Helsinki, Finland Sérusiaux, E., Goffinet, B., Miadlikowska, J. and Vitikainen, O. (2009). Taxonomy, phylogeny and biogeography of the the lichen genus Peltigera in Papua New Guinea. Fungal Diversity 38: 185-224. The lichen genus Peltigera is represented in Papua New Guinea by 15 species, including 6 described as new for science: P. cichoracea, P. didactyla, P. dolichorhiza, P. erioderma, P. extenuata, P. fimbriata sp. nov., P. granulosa sp. nov., P. koponenii sp. nov., P. montis-wilhelmii sp. nov., P. nana, P. oceanica, P. papuana sp. nov., P. sumatrana, P. ulcerata, and P. weberi sp. nov. Peltigera macra and P. tereziana var. philippinensis are reduced to synonymy with P. nana, whereas P. melanocoma is maintained as a species distinct from P. nana pending further studies. The status of several putative taxa referred to P. dolichorhiza s. lat. in the Sect. Polydactylon remains to be studied on a wider geographical scale and in the context of P. dolichorhiza and P. neopolydactyla. The phylogenetic affinities of all but one regional species (P. extenuata) are studied based on inferences from ITS (nrDNA) sequence data, in the context of a broad taxonomic sampling within the genus. -

Boreal Felt Lichen (Erioderma Pedicellatum) Is a Globally Threatened, Conspicuous Foliose Cyanolichen Belonging to the Pannariaceae
COSEWIC Assessment and Status Report on the Boreal Felt Lichen Erioderma pedicellatum Atlantic population Boreal population in Canada ENDANGERED - Atlantic population 2002 SPECIAL CONCERN - Boreal population 2002 COSEWIC COSEPAC COMMITTEE ON THE STATUS OF COMITÉ SUR LA SITUATION DES ENDANGERED WILDLIFE IN ESPÈCES EN PÉRIL CANADA AU CANADA COSEWIC status reports are working documents used in assigning the status of wildlife species suspected of being at risk. This report may be cited as follows: Please note: Persons wishing to cite data in the report should refer to the report (and cite the author(s)); persons wishing to cite the COSEWIC status will refer to the assessment (and cite COSEWIC). A production note will be provided if additional information on the status report history is required. COSEWIC 2002. COSEWIC assessment and status report on the boreal felt lichen Erioderma pedicellatum in Canada. Committee on the Status of Endangered Wildlife in Canada. Ottawa. viii + 50 pp. Maass, W. and D. Yetman. 2002. COSEWIC assessment and status report on the boreal felt lichen Erioderma pedicellatum in Canada, in COSEWIC assessment and status report on the boreal felt lichen Erioderma pedicellatum in Canada. Committee on the Status of Endangered Wildlife in Canada. Ottawa. 1- 50 pp. For additional copies contact: COSEWIC Secretariat c/o Canadian Wildlife Service Environment Canada Ottawa, ON K1A 0H3 Tel.: (819) 997-4991 / (819) 953-3215 Fax: (819) 994-3684 E-mail: COSEWIC/[email protected] http://www.cosewic.gc.ca Ếgalement disponible en français sous le titre Évaluation et Rapport du COSEPAC sur la situation de l’erioderme boréal (Erioderma pedicellatum) au Canada Cover illustration: Boreal felt lichen — Provided by the author, photo by Dr. -

Piedmont Lichen Inventory
PIEDMONT LICHEN INVENTORY: BUILDING A LICHEN BIODIVERSITY BASELINE FOR THE PIEDMONT ECOREGION OF NORTH CAROLINA, USA By Gary B. Perlmutter B.S. Zoology, Humboldt State University, Arcata, CA 1991 A Thesis Submitted to the Staff of The North Carolina Botanical Garden University of North Carolina at Chapel Hill Advisor: Dr. Johnny Randall As Partial Fulfilment of the Requirements For the Certificate in Native Plant Studies 15 May 2009 Perlmutter – Piedmont Lichen Inventory Page 2 This Final Project, whose results are reported herein with sections also published in the scientific literature, is dedicated to Daniel G. Perlmutter, who urged that I return to academia. And to Theresa, Nichole and Dakota, for putting up with my passion in lichenology, which brought them from southern California to the Traingle of North Carolina. TABLE OF CONTENTS Introduction……………………………………………………………………………………….4 Chapter I: The North Carolina Lichen Checklist…………………………………………………7 Chapter II: Herbarium Surveys and Initiation of a New Lichen Collection in the University of North Carolina Herbarium (NCU)………………………………………………………..9 Chapter III: Preparatory Field Surveys I: Battle Park and Rock Cliff Farm……………………13 Chapter IV: Preparatory Field Surveys II: State Park Forays…………………………………..17 Chapter V: Lichen Biota of Mason Farm Biological Reserve………………………………….19 Chapter VI: Additional Piedmont Lichen Surveys: Uwharrie Mountains…………………...…22 Chapter VII: A Revised Lichen Inventory of North Carolina Piedmont …..…………………...23 Acknowledgements……………………………………………………………………………..72 Appendices………………………………………………………………………………….…..73 Perlmutter – Piedmont Lichen Inventory Page 4 INTRODUCTION Lichens are composite organisms, consisting of a fungus (the mycobiont) and a photosynthesising alga and/or cyanobacterium (the photobiont), which together make a life form that is distinct from either partner in isolation (Brodo et al. -

Cyanobacteria Produce a High Variety of Hepatotoxic Peptides in Lichen Symbiosis
Cyanobacteria produce a high variety of hepatotoxic peptides in lichen symbiosis Ulla Kaasalainena,1, David P. Fewerb, Jouni Jokelab, Matti Wahlstenb, Kaarina Sivonenb, and Jouko Rikkinena aDepartment of Biosciences and bDepartment of Food and Environmental Sciences, Division of Microbiology, University of Helsinki, FIN-00014, Helsinki, Finland Edited by Robert Haselkorn, University of Chicago, Chicago, IL, and approved February 28, 2012 (received for review January 6, 2012) Lichens are symbiotic associations between fungi and photosyn- freshwater ecosystems. We have previously shown that the Nostoc thetic algae or cyanobacteria. Microcystins are potent toxins that symbionts of the tripartite cyanolichen species Peltigera leuco- are responsible for the poisoning of both humans and animals. phlebia can produce hepatotoxic microcystins in lichen symbiosis These toxins are mainly associated with aquatic cyanobacterial (11). However, it was unclear whether the production of these blooms, but here we show that the cyanobacterial symbionts of potent hepatotoxins in lichen symbiosis is a frequent phenome- terrestrial lichens from all over the world commonly produce non. Here we report that microcystins and nodularins are pro- microcystins. We screened 803 lichen specimens from five different duced in many different cyanolichen lineages and climatic regions continents for cyanobacterial toxins by amplifying a part of the all over the world. gene cluster encoding the enzyme complex responsible for micro- cystin production and detecting toxins directly from lichen thalli. We Results found either the biosynthetic genes for making microcystins or the A total of 803 lichen thalli representing 23 different cyanolichen toxin itself in 12% of all analyzed lichen specimens. A plethora of genera from different parts of the world were analyzed (Fig. -

Medicinal Lichens: the Final Frontier by Brian Kie Weissbuch, L.Ac., RH (AHG)
J A H G Volume 12 | Number 2 Journal of the American Herbalists Guild 23 MATERIA MEDICA MATERIA Medicinal Lichens: The Final Frontier by Brian Kie Weissbuch, L.Ac., RH (AHG) erbalists of myriad traditions article discusses recent scienti!c research Brian Kie Weissbuch L.Ac., AHG, is an acupuncturist have explored the healing indicating important uses for well-known lichens in private practice since properties of life-forms from at (Cetraria islandica and Usnea spp.) as well as less 1991. Brian is a botanist least !ve kingdoms: Monera familiar species (Flavoparmelia caperata and and herbalist with 44 years (bacteria)*, Protista (algae), Lobaria pulmonaria ). experience, and is co- Fungi, Plantae, and Animalia *Recall the ancient Nubians’ crafting of founder of KW Botanicals H Inc. in San Anselmo, CA, (the latter being primarily the domain of TCM medicinal beers, rich in tetracycline from the providing medicinal herb practitioners, who use gecko, snake, oyster shell, soil-borne Streptomyces bacteria found on the brewers’ formulae to primary health various insects, and other animal-derived grain (Nelson et al 2010). This medicine was given to care practitioners since 1983. substances). Nevertheless, with few exceptions, children as well as adults. He provides continuing herbalists have neglected an important life-form in education seminars to our medicines: the symbiotic intersection of algae Cetraria islandica, C. spp. The Iceland acupuncturists, and classes in herbal medicine for herbalists and fungi with a history of over 600 million years Mosses and health care practitioners. on our wayward planet —the lichens. Any discussion of edible and medicinal lichens He can be reached at brian@ How little we know of this symbiotic life- must begin with Cetraria islandica and allied kwbotanicals.com. -

Distribution and National Conservation Status of the Lichen
Mycosphere 8(4): 630–648 (2017) www.mycosphere.org ISSN 2077 7019 Article Doi 10.5943/mycosphere/8/4/10 Copyright © Guizhou Academy of Agricultural Sciences Distribution and national conservation status of the lichen family Lobariaceae (Peltigerales): from subtropical luxuriant forests to the alpine scrub of Nepal Himalaya Devkota S1*, Keller C1, Olley L2, Werth S1,3, Chaudhary RP4 and Scheidegger C1 1 Swiss Federal Research Institute WSL, CH-8903 Birmensdorf, Switzerland 2 Royal Botanic Gardens, Edinburgh (RBGE) EH3 5LR, Scotland, UK 3 Institute of Plant Sciences, University of Graz, 8010 Graz, Austria 4Research Centre for Applied Science and Technology (RECAST), Tribhuvan University, Kirtipur, Nepal Devkota S, Keller C, Olley L, Werth S, Chaudhary RP, Scheidegger C 2017 – Distribution and national conservation status of the lichen family Lobariaceae (Peltigerales): from subtropical luxuriant forests to the alpine scrub of Nepal Himalaya. Mycosphere 8(4), 630–648, Doi 10.5943/mycosphere/8/4/10 Abstract During 2007 - 2014, voucher specimens of Lobariaceae were collected from different geographic locations of Taplejung, Solukhumbu, Rasuwa, Gorkha, Manang, Kaski, and Myagdi districts of Nepal. Morphological characters, chemical tests and thin-layer chromatography techniques (TLC) were applied for the identification. Combining with earlier publications on Lobariaceae, this study summarized two genera Lobaria and Sticta each with seven and six species, reported from ten different districts of Nepal. The altitudinal distribution of the species varies from 1350 m to 5004 m (i.e. subtropical to alpine bioclimatic zones) above sea level, from Eastern, Central and Western parts of Nepal. Lobaria adscripturiens (Nyl.) Hue, L. fuscotomentosa Yoshim. L. aff. -
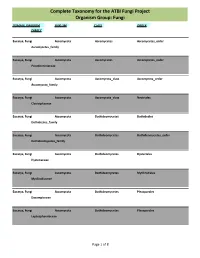
Complete Taxonomy for the ATBI Fungi Project Organism Group: Fungi
Complete Taxonomy for the ATBI Fungi Project Organism Group: Fungi DOMAIN, KINGDOM PHYLUM CLASS ORDER FAMILY Eucarya, Fungi Ascomycota Ascomycetes Ascomycetes_order Ascomycetes_family Eucarya, Fungi Ascomycota Ascomycetes Ascomycetes_order Pseudeurotiaceae Eucarya, Fungi Ascomycota Ascomycota_class Ascomycota_order Ascomycota_family Eucarya, Fungi Ascomycota Ascomycota_class Nectriales Clavicipitaceae Eucarya, Fungi Ascomycota Dothideomycetes Dothideales Dothideales_family Eucarya, Fungi Ascomycota Dothideomycetes Dothideomycetes_order Dothideomycetes_family Eucarya, Fungi Ascomycota Dothideomycetes Hysteriales Hysteriaceae Eucarya, Fungi Ascomycota Dothideomycetes Mytilinidiales Mytilinidiaceae Eucarya, Fungi Ascomycota Dothideomycetes Pleosporales Dacampiaceae Eucarya, Fungi Ascomycota Dothideomycetes Pleosporales Leptosphaeriaceae Page 1 of 8 Complete Taxonomy for the ATBI Fungi Project Organism Group: Fungi DOMAIN, KINGDOM PHYLUM CLASS ORDER FAMILY Eucarya, Fungi Ascomycota Dothideomycetes Pleosporales Lophiostomataceae Eucarya, Fungi Ascomycota Dothideomycetes Pleosporales Massarinaceae Eucarya, Fungi Ascomycota Dothideomycetes Pleosporales Melanommataceae Eucarya, Fungi Ascomycota Dothideomycetes Pleosporales Pleomassariaceae Eucarya, Fungi Ascomycota Dothideomycetes Pleosporales Pleosporaceae Eucarya, Fungi Ascomycota Dothideomycetes Pleosporales Pleosporales Eucarya, Fungi Ascomycota Dothideomycetes Pleosporales Pleosporales_family Eucarya, Fungi Ascomycota Dothideomycetes Pleosporales Sporormiaceae Eucarya, Fungi Ascomycota Dothideomycetes -

Diversity and Distribution Patterns of Endolichenic Fungi in Jeju Island, South Korea
sustainability Article Diversity and Distribution Patterns of Endolichenic Fungi in Jeju Island, South Korea Seung-Yoon Oh 1,2 , Ji Ho Yang 1, Jung-Jae Woo 1,3, Soon-Ok Oh 3 and Jae-Seoun Hur 1,* 1 Korean Lichen Research Institute, Sunchon National University, 255 Jungang-Ro, Suncheon 57922, Korea; [email protected] (S.-Y.O.); [email protected] (J.H.Y.); [email protected] (J.-J.W.) 2 Department of Biology and Chemistry, Changwon National University, 20 Changwondaehak-ro, Changwon 51140, Korea 3 Division of Forest Biodiversity, Korea National Arboretum, 415 Gwangneungsumok-ro, Pocheon 11186, Korea; [email protected] * Correspondence: [email protected]; Tel.: +82-61-750-3383 Received: 24 March 2020; Accepted: 1 May 2020; Published: 6 May 2020 Abstract: Lichens are symbiotic organisms containing diverse microorganisms. Endolichenic fungi (ELF) are one of the inhabitants living in lichen thalli, and have potential ecological and industrial applications due to their various secondary metabolites. As the function of endophytic fungi on the plant ecology and ecosystem sustainability, ELF may have an influence on the lichen diversity and the ecosystem, functioning similarly to the influence of endophytic fungi on plant ecology and ecosystem sustainability, which suggests the importance of understanding the diversity and community pattern of ELF. In this study, we investigated the diversity and the factors influencing the community structure of ELF in Jeju Island, South Korea by analyzing 619 fungal isolates from 79 lichen samples in Jeju Island. A total of 112 ELF species was identified and the most common species belonged to Xylariales in Sordariomycetes. -

BOSCASTLE to WIDEMOUTH DISTRICT: North Cornwall Status
COUNTY: Cornwall SITE NAME: BOSCASTLE TO WIDEMOUTH DISTRICT: North Cornwall Status: Site of Special Scientific Interest (SSSI) notified under Section 28 of the Wildlife and Countryside Act 1981 (as amended) Local Planning Authority: Cornwall County Council, North Cornwall District Council National Grid Reference: SX 092916–SS 194018 Area: 639 (ha) 1579 (ac) Ordnance Survey Sheet 1:50,000: 190 1:10,000: SX 09 SE, SX 19 SW, NW, NE, SS 10 SE Date Notified (Under 1949 Act): 1972 Date of Last Revision: – Date Notified (Under 1981 Act): 1990 Date of Last Revision: – Other Information: Within an Area of Outstanding Natural Beauty and North Cornwall Heritage Coast; part owned by the National Trust; includes 5 Geological Conservation Review sites and is noted in “A Nature Conservation Review” Ed. D A Ratcliffe (Cambridge University Press 1977). Site amended by extensions and deletions. Description and Reasons for Notification: This site lies on the North Cornwall coast and comprises a 12 mile section of cliffs and coastal habitats between Boscastle and Widemouth. The cliffs exhibit classic geological exposures of Namurian rocks and Variscan structures; the outstanding biological interest includes the unique Dizzard Oak woodland, maritime heaths and intertidal zones. Five Geological Conservation Review Localities occur within the site: 1. Widemouth to Crackington – This site is comprised of extensive coastal exposures, where the typically developed basinal Namurian of south-west England is clearly exposed. The entire Namurian represented by the Crackington formation is visible within the site, and the presence of rare goniatites has been vital in unravelling the complicated local stratigraphy. The section provides an excellent display of the sedimentary features associated with shallow water turbidites, and is of considerable interest for its spectacular structural features.