TRACE Document: Erickson Et Al
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Metro Response to Public Comments Proposed License for Grimm's Fuel
Metro response to public comments Proposed license for Grimm’s Fuel Company Metro response to public comments received during the comment period in October through November 2018 regarding proposed license conditions for Grimm’s Fuel Company. Prepared by Hila Ritter January 4, 2019 BACKGROUND Grimm’s Fuel Company (Grimm’s), is a locally-owned and operated Metro-licensed yard debris composting facility that primarily accepts yard debris for composting. In general, Metro’s licensing requirements for a composting facility set conditions for receiving, managing, and transferring yard debris and other waste at the facility. The Oregon Department of Environmental Quality (DEQ) also has distinct yet complementary requirements for composting facilities and monitors environmental impacts of facilities to protect air, soil, and water quality. Metro and DEQ work closely together to provide regulatory oversight of solid waste facilities such as Grimm’s. On October 22, 2018, Metro opened a formal public comment period for several new conditions of the draft license for Grimm’s. The public comment period closed on November 30, 2018. Metro also hosted a community conversation during that time to further solicit input and discuss next steps. During the formal comment period, Metro received 119 written comments from individuals and businesses (attached). Metro received an additional six written comments via email after the comment period closed at 5 p.m. on November 30, which did not become an official part of the record. After the comment period closed, members of the Tualatin community requested that Metro extend the term of Grimm’s current license for an additional two months to allow the public an opportunity to review the new proposed license in its entirety before it is issued. -

Mothers Grimm Kindle
MOTHERS GRIMM PDF, EPUB, EBOOK Danielle Wood | 224 pages | 01 Oct 2016 | Allen & Unwin | 9781741756746 | English | St Leonards, Australia Mothers Grimm PDF Book Showing An aquatic reptilian-like creature that is an exceptional swimmer. They have a temper that they control and release to become effective killers, particularly when a matter involves a family member or loved one. She took Nick to Weston's car and told Nick that he knew Adalind was upstairs with Renard, and the two guys Weston sent around back knew too. When Wu asks how she got over thinking it was real, she tells him that it didn't matter whether it was real, what mattered was losing her fear of it. Dick Award Nominee I found the characters appealing, and the plot intriguing. This wesen is portrayed as the mythological basis for the Three Little Pigs. The tales are very dark, and while the central theme is motherhood, the stories are truly about womanhood, and society's unrealistic and unfair expectations of all of us. Paperback , pages. The series presents them as the mythological basis for The Story of the Three Bears. In a phone call, his parents called him Monroe, seeming to indicate that it is his first name. The first edition contained 86 stories, and by the seventh edition in , had unique fairy tales. Danielle is currently teaching creative writing at the University of Tasmania. The kiss of a musai secretes a psychotropic substance that causes obsessive infatuation. View all 3 comments. He asks Sean Renard, a police captain, to endorse him so he would be elected for the mayor position. -
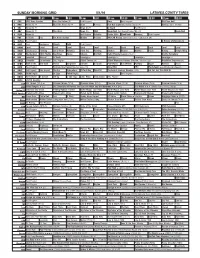
Sunday Morning Grid 5/1/16 Latimes.Com/Tv Times
SUNDAY MORNING GRID 5/1/16 LATIMES.COM/TV TIMES 7 am 7:30 8 am 8:30 9 am 9:30 10 am 10:30 11 am 11:30 12 pm 12:30 2 CBS CBS News Sunday Face the Nation (N) Paid Program Boss Paid Program PGA Tour Golf 4 NBC News (N) Å Meet the Press (N) Å News Rescue Red Bull Signature Series (Taped) Å Hockey: Blues at Stars 5 CW News (N) Å News (N) Å In Touch Paid Program 7 ABC News (N) Å This Week News (N) NBA Basketball First Round: Teams TBA. (N) Basketball 9 KCAL News (N) Joel Osteen Schuller Pastor Mike Woodlands Amazing Paid Program 11 FOX In Touch Paid Fox News Sunday Midday Prerace NASCAR Racing Sprint Cup Series: GEICO 500. (N) 13 MyNet Paid Program A History of Violence (R) 18 KSCI Paid Hormones Church Faith Paid Program 22 KWHY Local Local Local Local Local Local Local Local Local Local Local Local 24 KVCR Landscapes Painting Joy of Paint Wyland’s Paint This Painting Kitchen Mexico Martha Pépin Baking Simply Ming 28 KCET Wunderkind 1001 Nights Bug Bites Space Edisons Biz Kid$ Celtic Thunder Legacy (TVG) Å Soulful Symphony 30 ION Jeremiah Youssef In Touch Leverage Å Leverage Å Leverage Å Leverage Å 34 KMEX Conexión En contacto Paid Program Fútbol Central (N) Fútbol Mexicano Primera División: Toluca vs Azul República Deportiva (N) 40 KTBN Walk in the Win Walk Prince Carpenter Schuller In Touch PowerPoint It Is Written Pathway Super Kelinda Jesse 46 KFTR Paid Program Formula One Racing Russian Grand Prix. -

Download Season 5 Grimm Free Watch Grimm Season 5 Episode 1 Online
download season 5 grimm free Watch Grimm Season 5 Episode 1 Online. On Grimm Season 5 Episode 1, Nick struggles with how to move forward after the beheading of his mother, the death of Juliette and Adalind being pregnant. Watch Similar Shows FREE Amazon Watch Now iTunes Watch Now Vudu Watch Now YouTube Purchase Watch Now Google Play Watch Now NBC Watch Now Verizon On Demand Watch Now. When you watch Grimm Season 5 Episode 1 online, the action picks up right where the season finale of Grimm Season 4 left off, with Juliette dead in Nick's arms, Nick's mother's severed head in a box, and Agent Chavez's goons coming to kidnap Trubel. Nick is apparently drugged, and when he awakes, there is no sign that any of this occurred: no body, no box, no head, no Trubel. Was it all a truly horrible dream? Or part of a larger conspiracy? When he tries to tell his friends what happened, they hesitate to believe his claims, especially when he lays the blame at the feet of Agent Chavez. As his mental state deteriorates, will Nick be able to discover the truth about what happened and why? Where is Trubel now, and why did Chavez and her people come to kidnap her? Will his friends ever believe him, or will they continue to doubt his claims about what happened? And what will Nick do now that his and Adalind's baby is on the way? Tune in and watch Grimm Season 5 Episode 1, "The Grimm Identity," online right here at TV Fanatic to find out what happens. -

El Llegat Dels Germans Grimm En El Segle Xxi: Del Paper a La Pantalla Emili Samper Prunera Universitat Rovira I Virgili [email protected]
El llegat dels germans Grimm en el segle xxi: del paper a la pantalla Emili Samper Prunera Universitat Rovira i Virgili [email protected] Resum Les rondalles que els germans Grimm van recollir als Kinder- und Hausmärchen han traspassat la frontera del paper amb nombroses adaptacions literàries, cinema- togràfiques o televisives. La pel·lícula The brothers Grimm (2005), de Terry Gilli- am, i la primera temporada de la sèrie Grimm (2011-2012), de la cadena NBC, són dos mostres recents d’obres audiovisuals que han agafat les rondalles dels Grimm com a base per elaborar la seva ficció. En aquest article s’analitza el tractament de les rondalles que apareixen en totes dues obres (tenint en compte un precedent de 1962, The wonderful world of the Brothers Grimm), així com el rol que adopten els mateixos germans Grimm, que passen de creadors a convertir-se ells mateixos en personatges de ficció. Es recorre, d’aquesta manera, el camí invers al que han realitzat els responsables d’aquestes adaptacions: de la pantalla (gran o petita) es torna al paper, mostrant quines són les rondalles dels Grimm que s’han adaptat i de quina manera s’ha dut a terme aquesta adaptació. Paraules clau Grimm, Kinder- und Hausmärchen, The brothers Grimm, Terry Gilliam, rondalla Summary The tales that the Grimm brothers collected in their Kinder- und Hausmärchen have gone beyond the confines of paper with numerous literary, cinematographic and TV adaptations. The film The Brothers Grimm (2005), by Terry Gilliam, and the first season of the series Grimm (2011–2012), produced by the NBC network, are two recent examples of audiovisual productions that have taken the Grimm brothers’ tales as a base on which to create their fiction. -

VR White-Paper.Indd
Virtual Reality at Grimm + Parker Architects Date: November 2, 2017 By: Antonio Rebelo AIA, Director of Design (Grimm + Parker Architects), in collaboration with John Schippers Contact: [email protected] 301.595.1000 Since the beginning of recorded design, the search for a method of conveying an idea of a three-dimensional space or environment that didn’t yet exist in physical form, encouraged the creation of perspective views, the perfection of hand painted renderings, sketches and even scaled down physical models throughout history. The practical necessity to “see” and even “feel” a three-dimensional space to test for proportion, light, materiality, context and a multitude of other elements, has been at the core of architecture practice since its inception. This need has been there to fulfi ll two basic requirements of the design process: to help the designer transform abstract ideas into form, by allowing for study through the iterative design process, and second, to off er others (colleagues, clients and the community) a means for the visualization of the designer’s intent and the client’s vision. With the advent of computer aided design and advance in technology, the profes- sion has seen a rapid evolution towards ever more realistic renderings and visualization in ways that were almost inconceivable in the past, such as full immersive virtual reality. From a client’s perspective, this evolution in technology couldn’t be better news! Instead Grimm + Parker Architects 11720 Beltsville Dr, Ste #600 of relying on imagination by trying to understand traditional 2D fl oor plans and elevations, Calverton, MD 20705 which can be notoriously diffi cult, even if these were paired with illustrations of interior Tel: 301.595.1000 and exterior spaces, virtual reality and full 3D design off er a much easier way to process Visit online at grimmandparker.com architectural information than ever before - for everyone. -

Anne Sexton and Sylvia Plath from a Kristevan Perspective
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by OpenGrey Repository Transforming the Law of One: Anne Sexton and Sylvia Plath from a Kristevan Perspective A thesis submitted for the degree of Doctor of Philosophy By Areen Ghazi Khalifeh School of Arts, Brunel University November 2010 ii Abstract A recent trend in the study of Anne Sexton and Sylvia Plath often dissociates Confessional poetry from the subject of the writer and her biography, claiming that the artist is in full control of her work and that her art does not have naïve mimetic qualities. However, this study proposes that subjective attributes, namely negativity and abjection, enable a powerful transformative dialectic. Specifically, it demonstrates that an emphasis on the subjective can help manifest the process of transgressing the law of One. The law of One asserts a patriarchal, monotheistic law as a social closed system and can be opposed to the bodily drives and its open dynamism. This project asserts that unique, creative voices are derived from that which is individual and personal and thus, readings of Confessional poetry are in fact best served by acknowledgment of the subjective. In order to stress the subject of the artist in Confessionalism, this study employed a psychoanalytical Kristevan approach. This enables consideration of the subject not only in terms of the straightforward narration of her life, but also in relation to her poetic language and the process of creativity where instinctual drives are at work. This study further applies a feminist reading to the subject‘s poetic language and its ability to transgress the law, not necessarily in the political, macrocosmic sense of the word, but rather on the microcosmic, subjective level. -
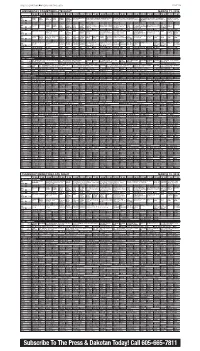
Subscribe to the Press & Dakotan Today!
PRESS & DAKOTAN n FRIDAY, MARCH 6, 2015 PAGE 9B WEDNESDAY PRIMETIME/LATE NIGHT MARCH 11, 2015 3:00 3:30 4:00 4:30 5:00 5:30 6:00 6:30 7:00 7:30 8:00 8:30 9:00 9:30 10:00 10:30 11:00 11:30 12:00 12:30 1:00 1:30 BROADCAST STATIONS Arthur Å Odd Wild Cyber- Martha Nightly PBS NewsHour (N) (In Suze Orman’s Financial Solutions for You 50 Years With Peter, Paul and Mary Perfor- John Sebastian Presents: Folk Rewind NOVA The tornado PBS (DVS) Squad Kratts Å chase Speaks Business Stereo) Å Finding financial solutions. (In Stereo) Å mances by Peter, Paul and Mary. (In Stereo) Å (My Music) Artists of the 1950s and ’60s. (In outbreak of 2011. (In KUSD ^ 8 ^ Report Stereo) Å Stereo) Å KTIV $ 4 $ Meredith Vieira Ellen DeGeneres News 4 News News 4 Ent Myst-Laura Law & Order: SVU Chicago PD News 4 Tonight Show Seth Meyers Daly News 4 Extra (N) Hot Bench Hot Bench Judge Judge KDLT NBC KDLT The Big The Mysteries of Law & Order: Special Chicago PD Two teen- KDLT The Tonight Show Late Night With Seth Last Call KDLT (Off Air) NBC (N) Å Å Judy Å Judy Å News Nightly News Bang Laura A star athlete is Victims Unit Å (DVS) age girls disappear. News Starring Jimmy Fallon Meyers (In Stereo) Å With Car- News Å KDLT % 5 % (N) Å News (N) (N) Å Theory killed. Å Å (DVS) (N) Å (In Stereo) Å son Daly KCAU ) 6 ) Dr. -

TLEX GRID (EAST REGULAR) - APRIL 2021 (4/12/2021 - 4/18/2021) - WEEK #16 Date Updated:3/25/2021 2:29:43 PM
TLEX GRID (EAST REGULAR) - APRIL 2021 (4/12/2021 - 4/18/2021) - WEEK #16 Date Updated:3/25/2021 2:29:43 PM MON (4/12/2021) TUE (4/13/2021) WED (4/14/2021) THU (4/15/2021) FRI (4/16/2021) SAT (4/17/2021) SUN (4/18/2021) SHOP LC (PAID PROGRAM SHOP LC (PAID PROGRAM SHOP LC (PAID PROGRAM SHOP LC (PAID PROGRAM SHOP LC (PAID PROGRAM SHOP LC (PAID PROGRAM SHOP LC (PAID PROGRAM 05:00A 05:00A NETWORK) NETWORK) NETWORK) NETWORK) NETWORK) NETWORK) NETWORK) PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM 05:30A 05:30A (NETWORK) (NETWORK) (NETWORK) (NETWORK) (NETWORK) (NETWORK) (NETWORK) PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM 06:00A 06:00A (NETWORK) (NETWORK) (NETWORK) (NETWORK) (NETWORK) (NETWORK) (NETWORK) PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM 06:30A 06:30A (SUBNETWORK) (SUBNETWORK) (SUBNETWORK) (SUBNETWORK) (SUBNETWORK) (NETWORK) (NETWORK) PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM 07:00A 07:00A (NETWORK) (NETWORK) (NETWORK) (NETWORK) (NETWORK) (NETWORK) (SUBNETWORK) PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM 07:30A 07:30A (NETWORK) (NETWORK) (NETWORK) (NETWORK) (NETWORK) (NETWORK) (SUBNETWORK) PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM 08:00A 08:00A (NETWORK) (NETWORK) (NETWORK) (NETWORK) (NETWORK) (NETWORK) (NETWORK) CASO CERRADO CASO CERRADO CASO CERRADO -

Eric Laneuville Филм ÑÐ ¿Ð¸ÑÑ ŠÐº (ФилмографиÑ)
Eric Laneuville Филм ÑÐ ¿Ð¸ÑÑ ŠÐº (ФилмографиÑ) Exposed https://bg.listvote.com/lists/film/movies/exposed-5421584/actors Maréchaussée https://bg.listvote.com/lists/film/movies/mar%C3%A9chauss%C3%A9e-26214860/actors Cry Lusion https://bg.listvote.com/lists/film/movies/cry-lusion-18644046/actors The Grimm Identity https://bg.listvote.com/lists/film/movies/the-grimm-identity-27070622/actors PTZD https://bg.listvote.com/lists/film/movies/ptzd-15930037/actors The Other Side https://bg.listvote.com/lists/film/movies/the-other-side-16639708/actors The Inheritance https://bg.listvote.com/lists/film/movies/the-inheritance-26025124/actors Key Move https://bg.listvote.com/lists/film/movies/key-move-29097907/actors The Other Woman https://bg.listvote.com/lists/film/movies/the-other-woman-1050294/actors Red Badge https://bg.listvote.com/lists/film/movies/red-badge-105095288/actors Red Herring https://bg.listvote.com/lists/film/movies/red-herring-105095322/actors Pink Chanel Suit https://bg.listvote.com/lists/film/movies/pink-chanel-suit-105095359/actors Like a Redheaded Stepchild https://bg.listvote.com/lists/film/movies/like-a-redheaded-stepchild-105095404/actors Little Red Book https://bg.listvote.com/lists/film/movies/little-red-book-105095422/actors The Redshirt https://bg.listvote.com/lists/film/movies/the-redshirt-105095436/actors Ruddy Cheeks https://bg.listvote.com/lists/film/movies/ruddy-cheeks-105095453/actors Days of Wine and Roses https://bg.listvote.com/lists/film/movies/days-of-wine-and-roses-105095500/actors Red Listed -

Regulation of Mammalian Hibernation. John Thomas Burns Louisiana State University and Agricultural & Mechanical College
Louisiana State University LSU Digital Commons LSU Historical Dissertations and Theses Graduate School 1977 Regulation of Mammalian Hibernation. John Thomas Burns Louisiana State University and Agricultural & Mechanical College Follow this and additional works at: https://digitalcommons.lsu.edu/gradschool_disstheses Recommended Citation Burns, John Thomas, "Regulation of Mammalian Hibernation." (1977). LSU Historical Dissertations and Theses. 3097. https://digitalcommons.lsu.edu/gradschool_disstheses/3097 This Dissertation is brought to you for free and open access by the Graduate School at LSU Digital Commons. It has been accepted for inclusion in LSU Historical Dissertations and Theses by an authorized administrator of LSU Digital Commons. For more information, please contact [email protected]. INFORMATION TO USERS Thii material was produced from a microfilm copy of the original document. While the most advanced technological means to photograph and reproduce this document have been used, the quality is heavily dependent upon the quality of the original submitted. The following explanation of techniques is provided to help you understand markings or patterns which may appear on this reproduction. 1.The sign or "target" for pages apparently lacking from the document photographed is "Missing Page(s)". If it was possible to obtain the missing page(s) or section, they are spliced into the film along with adjacent pages. This may have necessitated cutting thru an image and duplicating adjacent pages to insure you complete continuity. 2. When an imaga on the film is obliterated with a large round black mark, it is an indication that the photographer suspected that the copy may have moved during exposure and thus cause a blurred image. -

MAY 2021 (4/26/2021 - 5/2/2021) - WEEK #18 Date Updated:4/16/2021 2:24:45 PM
TLEX GRID (EAST REGULAR) - MAY 2021 (4/26/2021 - 5/2/2021) - WEEK #18 Date Updated:4/16/2021 2:24:45 PM MON (4/26/2021) TUE (4/27/2021) WED (4/28/2021) THU (4/29/2021) FRI (4/30/2021) SAT (5/1/2021) SUN (5/2/2021) SHOP LC (PAID PROGRAM SHOP LC (PAID PROGRAM SHOP LC (PAID PROGRAM SHOP LC (PAID PROGRAM SHOP LC (PAID PROGRAM SHOP LC (PAID PROGRAM SHOP LC (PAID PROGRAM 05:00A 05:00A NETWORK) NETWORK) NETWORK) NETWORK) NETWORK) NETWORK) NETWORK) PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM 05:30A 05:30A (NETWORK) (NETWORK) (NETWORK) (NETWORK) (NETWORK) (NETWORK) (NETWORK) PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM 06:00A 06:00A (NETWORK) (NETWORK) (NETWORK) (NETWORK) (NETWORK) (NETWORK) (NETWORK) PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM 06:30A 06:30A (SUBNETWORK) (SUBNETWORK) (SUBNETWORK) (SUBNETWORK) (SUBNETWORK) (NETWORK) (NETWORK) PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM 07:00A 07:00A (NETWORK) (NETWORK) (NETWORK) (NETWORK) (NETWORK) (NETWORK) (SUBNETWORK) PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM 07:30A 07:30A (NETWORK) (NETWORK) (NETWORK) (NETWORK) (NETWORK) (NETWORK) (SUBNETWORK) PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM PAID PROGRAM 08:00A 08:00A (NETWORK) (NETWORK) (NETWORK) (NETWORK) (NETWORK) (NETWORK) (NETWORK) CASO CERRADO CASO CERRADO CASO CERRADO CASO