Building Research Communities on Biological Pathways Thomas Kelder1,*, Martijn P
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Pathways and Networks Biological Meaning of the Gene Sets
Pathways and Networks Biological meaning of the gene sets Gene ontology terms ? Pathway mapping Linking to Pubmed abstracts or associted MESH terms Regulation by the same transcription factor (module) Protein families and domains Gene set enrichment analysis Over representation analysis 1 Gene set enrichment analysis 1. Given an a priori defined set of genes S. 2. Rank genes (e.g. by t‐value between 2 groups of microarray samples) ranked gene list L. 3. Calculation of an enrichment score (ES) that reflects the degree to which a set S is overrepresented at the extremes (top or bottom) of the entire ranked list L. 4. Estimation the statistical significance (nominal P value) of the ES by using an empirical phenotype‐based permutation test procedure. 5. Adjustment for multiple hypothesis testing by controlling the false discovery rate (FDR). Gene set enrichment analysis Subramanian A et al. Proc Natl Acad Sci (2005) 2 Biochemical and Metabolic Pathways Böhringer Mannheim Signaling networks (in cancer cell) Hannah, Weinberg, Cell. 2000 137 NCI curated pathway maps (http://pid.nci.nih.gov) Signal Transduction Knowledge Environment http://stke.sciencemag.org/cm/ 3 Pathways • Pathways are available mechanistic information •Kyoto Encylopedia of Genes and Genomes, KEGG (http://www.genome.jp/kegg/) BioCYc • EcoCYC Metabolic Pathway Database (E. Coli) •Also for other organisms (e.g. HumanCYC) 4 Pathways • Pathways from Biocarta • http://www.biocarta.com/genes/index.asp Transpath Part of larger BioBase package (commercial) • PathwayBuilder package -
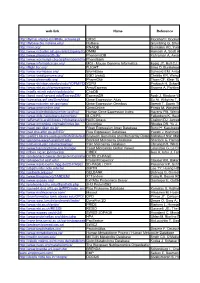
Web-Link Name Reference DRSC Flockhart I, Booker M, Kiger A, Et Al.: Flyrnai: the Drosophila Rnai Screening Center Database
web-link Name Reference http://flyrnai.org/cgi-bin/RNAi_screens.pl DRSC Flockhart I, Booker M, Kiger A, et al.: FlyRNAi: the Drosophila RNAi screening center database. Nucleic Acids Res. 34(Database issue):D489-94 (2006) http://flybase.bio.indiana.edu/ FlyBase Grumbling G, Strelets V.: FlyBase: anatomical data, images and queries. Nucleic Acids Res. 34(Database issue):D484-8 (2006) http://rnai.org/ RNAiDB Gunsalus KC, Yueh WC, MacMenamin P, et al.: RNAiDB and PhenoBlast: web tools for genome-wide phenotypic mapping projects. Nucleic Acids Res. 32(Database issue):D406-10 (2004) http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIMOMIM Hamosh A, Scott AF, Amberger JS, et al.: Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 33(Database issue):D514-7 (2005) http://www.phenomicdb.de/ PhenomicDB Kahraman A, Avramov A, Nashev LG, et al.: PhenomicDB: a multi-species genotype/phenotype database for comparative phenomics. Bioinformatics 21(3):418-20 (2005) http://www.worm.mpi-cbg.de/phenobank2/cgi-bin/MenuPage.pyPhenoBank http://www.informatics.jax.org/ MGI - Mouse Genome Informatics Eppig JT, Bult CJ, Kadin JA, et al.: The Mouse Genome Database (MGD): from genes to mice--a community resource for mouse biology. Nucleic Acids Res. 33(Database issue):D471-5 (2005) http://flight.licr.org/ FLIGHT Sims D, Bursteinas B, Gao Q, et al.: FLIGHT: database and tools for the integration and cross-correlation of large-scale RNAi phenotypic datasets. Nucleic Acids Res. 34(Database issue):D479-83 (2006) http://www.wormbase.org/ WormBase Schwarz EM, Antoshechkin I, Bastiani C, et al.: WormBase: better software, richer content. -

Global LC/MS Metabolomics Profiling of Calcium Stressed and Immunosuppressant Drug Treated Saccharomyces Cerevisiae
Metabolites 2013, 3, 1102-1117; doi:10.3390/metabo3041102 OPEN ACCESS metabolites ISSN 2218-1989 www.mdpi.com/journal/metabolites/ Article Global LC/MS Metabolomics Profiling of Calcium Stressed and Immunosuppressant Drug Treated Saccharomyces cerevisiae Stefan Jenkins 1,2,3,*, Steven M. Fischer 2, Lily Chen 3 and Theodore R. Sana 2 1 Life Sciences Division, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA 2 Agilent Technologies, Life Sciences, Diagnostics and Applied Markets, Santa Clara, CA 95051, USA; E-Mails: [email protected] (S.M.F.); [email protected] (T.R.S.) 3 Biology Department, San Francisco State University, San Francisco, CA 94132, USA; E-Mail: [email protected] * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +1-650-922-5046; Fax: +1-510-486-4545. Received: 15 October 2013; in revised form: 20 November 2013 / Accepted: 25 November 2013 / Published: 6 December 2013 Abstract: Previous studies have shown that calcium stressed Saccharomyces cerevisiae, challenged with immunosuppressant drugs FK506 and Cyclosporin A, responds with comprehensive gene expression changes and attenuation of the generalized calcium stress response. Here, we describe a global metabolomics workflow for investigating the utility of tracking corresponding phenotypic changes. This was achieved by efficiently analyzing relative abundance differences between intracellular metabolite pools from wild-type and calcium stressed cultures, with and without prior immunosuppressant drugs exposure. We used pathway database content from WikiPathways and YeastCyc to facilitate the projection of our metabolomics profiling results onto biological pathways. A key challenge was to increase the coverage of the detected metabolites. This was achieved by applying both reverse phase (RP) and aqueous normal phase (ANP) chromatographic separations, as well as electrospray ionization (ESI) and atmospheric pressure chemical ionization (APCI) sources for detection in both ion polarities. -

And Macro-Level Network Metrics Unveils Top Communicative Gene Modules in Psoriasis
G C A T T A C G G C A T genes Article Using Micro- and Macro-Level Network Metrics Unveils Top Communicative Gene Modules in Psoriasis 1, 2 3 Reyhaneh Naderi y, Homa Saadati Mollaei , Arne Elofsson 3, , and Saman Hosseini Ashtiani * y 1 Department of Artificial Intelligence and Robotics, Faculty of Computer Engineering, Iran University of Science and Technology, Tehran 1684613114, Iran; [email protected] 2 Department of Advanced Sciences and Technology, Islamic Azad University Tehran Medical Sciences, Tehran 1916893813, Iran; [email protected] 3 Department of Biochemistry and Biophysics and Science for Life Laboratory, Stockholm University, 106 91 Stockholm, Sweden; [email protected] * Correspondence: [email protected]; Tel.: +46-762623644 These authors contributed equally to this work and should be considered joint first authors. y Received: 9 July 2020; Accepted: 6 August 2020; Published: 10 August 2020 Abstract: (1) Background: Psoriasis is a multifactorial chronic inflammatory disorder of the skin, with significant morbidity, characterized by hyperproliferation of the epidermis. Even though psoriasis’ etiology is not fully understood, it is believed to be multifactorial, with numerous key components. (2) Methods: In order to cast light on the complex molecular interactions in psoriasis vulgaris at both protein–protein interactions and transcriptomics levels, we studied a set of microarray gene expression analyses consisting of 170 paired lesional and non-lesional samples. Afterwards, a network analysis was conducted on the protein–protein interaction network of differentially expressed genes based on micro- and macro-level network metrics at a systemic level standpoint. (3) Results: We found 17 top communicative genes, all of which were experimentally proven to be pivotal in psoriasis, which were identified in two modules, namely the cell cycle and immune system. -

Biological Pathways
Biological pathways Bing Zhang Department of Biomedical Informatics Vanderbilt University [email protected] Biological pathway A biological pathway is a series of actions among molecules in a cell that leads to a certain product or a change in a cell. Different types of biological pathways Signal transduction pathways Gene regulation pathways Metabolic pathways 2 Applied Bioinformatics, Spring 2011 Outline Pathway databases Pathway assembly and editing Pathway mapping and enrichment analysis 3 Applied Bioinformatics, Spring 2011 Selected pathway databases KEGG Kyoto Encyclopedia of Genes and Genomes http://www.genome.jp/kegg/pathway.html Reactome Ontario Institute for Cancer Research, Cold Spring Harbor Laboratory, New York University School of Medicine and The European Bioinformatics Institute http://www.reactome.org/ WikiPathways University of Maastricht and UCSF http://www.wikipathways.org/ 4 Applied Bioinformatics, Spring 2011 KEGG: data model Molecular building blocks KEGG GENES: genes and proteins generated by genome sequencing projects KEGG ORTHOLOGY: orthology (KO) groups KEGG COMPOUND: small molecules KEGG REACTIONS: biochemical reactions KEGG PATHWAY: pathway maps Created in a general way to be applicable to all organisms, in terms of the orthologs defined by KO groups Organism-specific pathways can be generated by converting KO groups to gene identifiers in a given organism 5 Applied Bioinformatics, Spring 2011 KEGG: TCA cycle 6 Applied Bioinformatics, Spring 2011 Reactome: data model Entities -

Genmapp 2: New Features and Resources for Pathway Analysis Nathan Salomonis Gladstone Institute of Cardiovascular Disease
Washington University School of Medicine Digital Commons@Becker Open Access Publications 2007 GenMAPP 2: New features and resources for pathway analysis Nathan Salomonis Gladstone Institute of Cardiovascular Disease Kristina Hanspers Gladstone Institute of Cardiovascular Disease Alexander C. Zambon Gladstone Institute of Cardiovascular Disease Karen Vranizan Gladstone Institute of Cardiovascular Disease Steven C. Lawlor Gladstone Institute of Cardiovascular Disease See next page for additional authors Follow this and additional works at: https://digitalcommons.wustl.edu/open_access_pubs Part of the Medicine and Health Sciences Commons Recommended Citation Salomonis, Nathan; Hanspers, Kristina; Zambon, Alexander C.; Vranizan, Karen; Lawlor, Steven C.; Dahlquist, Kam D.; Doniger, Scott .;W Stuart, Josh; Conklin, Bruce R.; and Pico, Alexander R., ,"GenMAPP 2: New features and resources for pathway analysis." BMC Bioinformatics.,. 217. (2007). https://digitalcommons.wustl.edu/open_access_pubs/208 This Open Access Publication is brought to you for free and open access by Digital Commons@Becker. It has been accepted for inclusion in Open Access Publications by an authorized administrator of Digital Commons@Becker. For more information, please contact [email protected]. Authors Nathan Salomonis, Kristina Hanspers, Alexander C. Zambon, Karen Vranizan, Steven C. Lawlor, Kam D. Dahlquist, Scott .W Doniger, Josh Stuart, Bruce R. Conklin, and Alexander R. Pico This open access publication is available at Digital Commons@Becker: https://digitalcommons.wustl.edu/open_access_pubs/208 -

Genmapp 2: New Features and Resources for Pathway Analysis Nathan Salomonis Gladstone Institute of Cardiovascular Disease
Washington University School of Medicine Digital Commons@Becker Open Access Publications 2007 GenMAPP 2: New features and resources for pathway analysis Nathan Salomonis Gladstone Institute of Cardiovascular Disease Kristina Hanspers Gladstone Institute of Cardiovascular Disease Alexander C. Zambon Gladstone Institute of Cardiovascular Disease Karen Vranizan Gladstone Institute of Cardiovascular Disease Steven C. Lawlor Gladstone Institute of Cardiovascular Disease See next page for additional authors Follow this and additional works at: http://digitalcommons.wustl.edu/open_access_pubs Part of the Medicine and Health Sciences Commons Recommended Citation Salomonis, Nathan; Hanspers, Kristina; Zambon, Alexander C.; Vranizan, Karen; Lawlor, Steven C.; Dahlquist, Kam D.; Doniger, Scott .;W Stuart, Josh; Conklin, Bruce R.; and Pico, Alexander R., ,"GenMAPP 2: New features and resources for pathway analysis." BMC Bioinformatics.8,. 217. (2007). http://digitalcommons.wustl.edu/open_access_pubs/208 This Open Access Publication is brought to you for free and open access by Digital Commons@Becker. It has been accepted for inclusion in Open Access Publications by an authorized administrator of Digital Commons@Becker. For more information, please contact [email protected]. Authors Nathan Salomonis, Kristina Hanspers, Alexander C. Zambon, Karen Vranizan, Steven C. Lawlor, Kam D. Dahlquist, Scott .W Doniger, Josh Stuart, Bruce R. Conklin, and Alexander R. Pico This open access publication is available at Digital Commons@Becker: http://digitalcommons.wustl.edu/open_access_pubs/208 -

Confirmation of Pathogenic Mechanisms by SARS-Cov-2–Host
Messina et al. Cell Death and Disease (2021) 12:788 https://doi.org/10.1038/s41419-021-03881-8 Cell Death & Disease ARTICLE Open Access Looking for pathways related to COVID-19: confirmation of pathogenic mechanisms by SARS-CoV-2–host interactome Francesco Messina 1, Emanuela Giombini1, Chiara Montaldo1, Ashish Arunkumar Sharma2, Antonio Zoccoli3, Rafick-Pierre Sekaly2, Franco Locatelli4, Alimuddin Zumla5, Markus Maeurer6,7, Maria R. Capobianchi1, Francesco Nicola Lauria1 and Giuseppe Ippolito 1 Abstract In the last months, many studies have clearly described several mechanisms of SARS-CoV-2 infection at cell and tissue level, but the mechanisms of interaction between host and SARS-CoV-2, determining the grade of COVID-19 severity, are still unknown. We provide a network analysis on protein–protein interactions (PPI) between viral and host proteins to better identify host biological responses, induced by both whole proteome of SARS-CoV-2 and specific viral proteins. A host-virus interactome was inferred, applying an explorative algorithm (Random Walk with Restart, RWR) triggered by 28 proteins of SARS-CoV-2. The analysis of PPI allowed to estimate the distribution of SARS-CoV-2 proteins in the host cell. Interactome built around one single viral protein allowed to define a different response, underlining as ORF8 and ORF3a modulated cardiovascular diseases and pro-inflammatory pathways, respectively. Finally, the network-based approach highlighted a possible direct action of ORF3a and NS7b to enhancing Bradykinin Storm. This network-based representation of SARS-CoV-2 infection could be a framework for pathogenic evaluation of specific 1234567890():,; 1234567890():,; 1234567890():,; 1234567890():,; clinical outcomes. -

Transcriptomic Uniqueness and Commonality of the Ion Channels and Transporters in the Four Heart Chambers Sanda Iacobas1, Bogdan Amuzescu2 & Dumitru A
www.nature.com/scientificreports OPEN Transcriptomic uniqueness and commonality of the ion channels and transporters in the four heart chambers Sanda Iacobas1, Bogdan Amuzescu2 & Dumitru A. Iacobas3,4* Myocardium transcriptomes of left and right atria and ventricles from four adult male C57Bl/6j mice were profled with Agilent microarrays to identify the diferences responsible for the distinct functional roles of the four heart chambers. Female mice were not investigated owing to their transcriptome dependence on the estrous cycle phase. Out of the quantifed 16,886 unigenes, 15.76% on the left side and 16.5% on the right side exhibited diferential expression between the atrium and the ventricle, while 5.8% of genes were diferently expressed between the two atria and only 1.2% between the two ventricles. The study revealed also chamber diferences in gene expression control and coordination. We analyzed ion channels and transporters, and genes within the cardiac muscle contraction, oxidative phosphorylation, glycolysis/gluconeogenesis, calcium and adrenergic signaling pathways. Interestingly, while expression of Ank2 oscillates in phase with all 27 quantifed binding partners in the left ventricle, the percentage of in-phase oscillating partners of Ank2 is 15% and 37% in the left and right atria and 74% in the right ventricle. The analysis indicated high interventricular synchrony of the ion channels expressions and the substantially lower synchrony between the two atria and between the atrium and the ventricle from the same side. Starting with crocodilians, the heart pumps the blood through the pulmonary circulation and the systemic cir- culation by the coordinated rhythmic contractions of its upper lef and right atria (LA, RA) and lower lef and right ventricles (LV, RV). -

1471-2105-8-217.Pdf
BMC Bioinformatics BioMed Central Software Open Access GenMAPP 2: new features and resources for pathway analysis Nathan Salomonis1,2, Kristina Hanspers1, Alexander C Zambon1, Karen Vranizan1,3, Steven C Lawlor1, Kam D Dahlquist4, Scott W Doniger5, Josh Stuart6, Bruce R Conklin1,2,7,8 and Alexander R Pico*1 Address: 1Gladstone Institute of Cardiovascular Disease, 1650 Owens Street, San Francisco, CA 94158 USA, 2Pharmaceutical Sciences and Pharmacogenomics Graduate Program, University of California, 513 Parnassus Avenue, San Francisco, CA 94143, USA, 3Functional Genomics Laboratory, University of California, Berkeley, CA 94720 USA, 4Department of Biology, Loyola Marymount University, 1 LMU Drive, MS 8220, Los Angeles, CA 90045 USA, 5Computational Biology Graduate Program, Washington University School of Medicine, St. Louis, MO 63108 USA, 6Department of Biomolecular Engineering, University of California, Santa Cruz, CA 95064 USA, 7Department of Medicine, University of California, San Francisco, CA 94143 USA and 8Department of Molecular and Cellular Pharmacology, University of California, San Francisco, CA 94143 USA Email: Nathan Salomonis - [email protected]; Kristina Hanspers - [email protected]; Alexander C Zambon - [email protected]; Karen Vranizan - [email protected]; Steven C Lawlor - [email protected]; Kam D Dahlquist - [email protected]; Scott W Doniger - [email protected]; Josh Stuart - [email protected]; Bruce R Conklin - [email protected]; Alexander R Pico* - [email protected] * Corresponding author Published: 24 June 2007 Received: 16 November 2006 Accepted: 24 June 2007 BMC Bioinformatics 2007, 8:217 doi:10.1186/1471-2105-8-217 This article is available from: http://www.biomedcentral.com/1471-2105/8/217 © 2007 Salomonis et al; licensee BioMed Central Ltd. -

Post-Graduate Diploma Program in Personalized Genomic Medicine” for Skill Development (DBT Sponsored Scheme)
UNIVERSITY OF MYSORE Syllabus for “Post-graduate Diploma Program in Personalized Genomic Medicine” For Skill Development (DBT Sponsored Scheme) FCBCS-CAGP SYSTEM Department of Studies in Genetics and Genomics University of Mysore Manasagangotri, Mysuru – 570 006 2019-20 1 PREAMBLE: The Information and potential use of genomic discoveries are no longer issues left for scientists and medical professionals to handle, but have become ones for the public at large. Rarely a day passes without genomics related story reported in the media. The proposed diploma program is designed to provide advanced knowledge dissemination in the field of genome sciences, applications and laboratory skills needed for molecular diagnostics and precision medicine procedures conducted in a clinical or research environment. The program is intended for those individuals who wish to enhance their laboratory expertise and knowledge in molecular-based methods. Precision medicine has the potential to fundamentally change how health care is practiced, but requires a trained health care workforce that understands the complexities of this field. One important component of precision medicine is the use of an individual’s genomic information to offer targeted treatment, tailored to the individual. Our course aims to provide participants with advanced knowledge of genomics, an overview of the clinical applications of genomic medicine, the skills to evaluate the clinical validity and utility of new tests, and an appreciation of the associated ethical and social issues inherent in this field. The course is geared towards individuals with a background in the biological sciences and a basic understanding of genetics. It is designed to be succinct and clinically focused, offering both conceptual and practical information about real-world applications of genomics. -

Chromatin Remodelling Complex Dosage Modulates Transcription Factor Function in Heart Development
ARTICLE Received 5 Aug 2010 | Accepted 11 Jan 2011 | Published 8 Feb 2011 DOI: 10.1038/ncomms1187 Chromatin remodelling complex dosage modulates transcription factor function in heart development Jun K. Takeuchi1,2,*, Xin Lou3,*, Jeffrey M. Alexander1,4, Hiroe Sugizaki2, Paul Delgado-Olguín1, Alisha K. Holloway1, Alessandro D. Mori1, John N. Wylie1, Chantilly Munson5,6, Yonghong Zhu3, Yu-Qing Zhou7, Ru-Fang Yeh8, R. Mark Henkelman7,9, Richard P. Harvey10,11, Daniel Metzger12, Pierre Chambon12, Didier Y. R. Stainier4,5,6, Katherine S. Pollard1,8, Ian C. Scott3,13 & Benoit G. Bruneau1,4,5,14 Dominant mutations in cardiac transcription factor genes cause human inherited congenital heart defects (CHDs); however, their molecular basis is not understood. Interactions between transcription factors and the Brg1/Brm-associated factor (BAF) chromatin remodelling complex suggest potential mechanisms; however, the role of BAF complexes in cardiogenesis is not known. In this study, we show that dosage of Brg1 is critical for mouse and zebrafish cardiogenesis. Disrupting the balance between Brg1 and disease-causing cardiac transcription factors, including Tbx5, Tbx20 and Nkx2–5, causes severe cardiac anomalies, revealing an essential allelic balance between Brg1 and these cardiac transcription factor genes. This suggests that the relative levels of transcription factors and BAF complexes are important for heart development, which is supported by reduced occupancy of Brg1 at cardiac gene promoters in Tbx5 haploinsufficient hearts. Our results reveal complex dosage-sensitive interdependence between transcription factors and BAF complexes, providing a potential mechanism underlying transcription factor haploinsufficiency, with implications for multigenic inheritance of CHDs. 1 Gladstone Institute of Cardiovascular Disease, San Francisco, California 94158, USA.