Dna Barcoding of Chlorarachniophytes Using Nucleomorph Its Sequences1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Molecular Data and the Evolutionary History of Dinoflagellates by Juan Fernando Saldarriaga Echavarria Diplom, Ruprecht-Karls-Un
Molecular data and the evolutionary history of dinoflagellates by Juan Fernando Saldarriaga Echavarria Diplom, Ruprecht-Karls-Universitat Heidelberg, 1993 A THESIS SUBMITTED IN PARTIAL FULFILMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY in THE FACULTY OF GRADUATE STUDIES Department of Botany We accept this thesis as conforming to the required standard THE UNIVERSITY OF BRITISH COLUMBIA November 2003 © Juan Fernando Saldarriaga Echavarria, 2003 ABSTRACT New sequences of ribosomal and protein genes were combined with available morphological and paleontological data to produce a phylogenetic framework for dinoflagellates. The evolutionary history of some of the major morphological features of the group was then investigated in the light of that framework. Phylogenetic trees of dinoflagellates based on the small subunit ribosomal RNA gene (SSU) are generally poorly resolved but include many well- supported clades, and while combined analyses of SSU and LSU (large subunit ribosomal RNA) improve the support for several nodes, they are still generally unsatisfactory. Protein-gene based trees lack the degree of species representation necessary for meaningful in-group phylogenetic analyses, but do provide important insights to the phylogenetic position of dinoflagellates as a whole and on the identity of their close relatives. Molecular data agree with paleontology in suggesting an early evolutionary radiation of the group, but whereas paleontological data include only taxa with fossilizable cysts, the new data examined here establish that this radiation event included all dinokaryotic lineages, including athecate forms. Plastids were lost and replaced many times in dinoflagellates, a situation entirely unique for this group. Histones could well have been lost earlier in the lineage than previously assumed. -

The Apicoplast: a Review of the Derived Plastid of Apicomplexan Parasites
Curr. Issues Mol. Biol. 7: 57-80. Online journalThe Apicoplastat www.cimb.org 57 The Apicoplast: A Review of the Derived Plastid of Apicomplexan Parasites Ross F. Waller1 and Geoffrey I. McFadden2,* way to apicoplast discovery with studies of extra- chromosomal DNAs recovered from isopycnic density 1Botany, University of British Columbia, 3529-6270 gradient fractionation of total Plasmodium DNA. This University Boulevard, Vancouver, BC, V6T 1Z4, Canada group recovered two DNA forms; one a 6kb tandemly 2Plant Cell Biology Research Centre, Botany, University repeated element that was later identifed as the of Melbourne, 3010, Australia mitochondrial genome, and a second, 35kb circle that was supposed to represent the DNA circles previously observed by microscopists (Wilson et al., 1996b; Wilson Abstract and Williamson, 1997). This molecule was also thought The apicoplast is a plastid organelle, homologous to to be mitochondrial DNA, and early sequence data of chloroplasts of plants, that is found in apicomplexan eubacterial-like rRNA genes supported this organellar parasites such as the causative agents of Malaria conclusion. However, as the sequencing effort continued Plasmodium spp. It occurs throughout the Apicomplexa a new conclusion, that was originally embraced with and is an ancient feature of this group acquired by the some awkwardness (“Have malaria parasites three process of endosymbiosis. Like plant chloroplasts, genomes?”, Wilson et al., 1991), began to emerge. apicoplasts are semi-autonomous with their own genome Gradually, evermore convincing character traits of a and expression machinery. In addition, apicoplasts import plastid genome were uncovered, and strong parallels numerous proteins encoded by nuclear genes. These with plastid genomes from non-photosynthetic plants nuclear genes largely derive from the endosymbiont (Epifagus virginiana) and algae (Astasia longa) became through a process of intracellular gene relocation. -

Norrisiella Sphaerica Gen
Norrisiella sphaerica gen. et sp. nov., a new coccoid chlorarachniophyte from Baja California, Mexico 著者 Ota Shuhei, Ueda Kunihiko, Ishida Ken-ichiro journal or Journal of Plant Research publication title volume 120 number 6 page range 661-670 year 2007-11-01 URL http://hdl.handle.net/2297/7674 doi: 10.1007/s10265-007-0115-y Norrisiella sphaerica gen. et sp. nov., a new coccoid chlorarachniophyte from Baja California, Mexico Shuhei Ota1, 2, Kunihiko Ueda1 and Ken-ichiro Ishida2 1Division of Life Sciences, Graduate School of Natural Science and Technology, Kanazawa University, Kakuma, Kanazawa 920-1192, Japan 2Graduate School of Life and Environmental Sciences, University of Tsukuba, 1-1-1 Tennodai, Tsukuba 305-8572, Japan Running title: Norrisiella sphaerica gen. et sp. nov. Correspondence: Shuhei Ota Laboratory of Plant Systematics and Phylogeny (D508) Institute of Biological Sciences Graduate School of Life and Environmental Sciences University of Tsukuba, 1-1-1, Tennodai, Tsukuba 305-8572, Japan Tel/Fax: +81 (29) 853-7267 Fax: +81 (29) 853-4533 e-mail: [email protected] 1/29 Abstract A new chlorarachniophyte, Norrisiella sphaerica S. Ota et K. Ishida gen. et sp. nov., is described from the coast of Baja California, Mexico. We examined its morphology, ultrastructure and life cycle in detail, using light microscopy, transmission electron microscopy and time-lapse videomicroscopy. We found that this chlorarachniophyte possessed the following characteristics: (i) vegetative cells were coccoid and possessed a cell wall, (ii) a pyrenoid was slightly invaded by plate-like periplastidial compartment from the tip of the pyrenoid, (iii) a nucleomorph was located near the pyrenoid base in the periplastidial compartment, (iv) cells reproduced vegetatively via autospores, and (v) a flagellate stage was present in the life cycle. -

Provirophages in the Bigelowiella Genome Bear Testimony to Past Encounters with Giant Viruses
Provirophages in the Bigelowiella genome bear testimony to past encounters with giant viruses Guillaume Blanca,1,2, Lucie Gallot-Lavalléea, and Florian Maumusb,1,2 aLaboratoire Information Génomique et Structurale, UMR7256 (Institut de Microbiologie de la Méditerranée FR3479) CNRS, Aix-Marseille Université, 13288 Marseille cedex 9, France; and bINRA, UR1164 Unité de Recherche Génomique-Info, Institut National de la Recherche Agronomique de Versailles-Grignon, 78026 Versailles, France Edited by Peter Palese, Icahn School of Medicine at Mount Sinai, New York, NY, and approved July 24, 2015 (received for review April 1, 2015) Virophages are recently discovered double-stranded DNA virus satel- cysteine protease (PRO), and zinc-ribbon domain (ZnR) as well as lites that prey on giant viruses (nucleocytoplasmic large DNA viruses; major and minor capsid proteins (MCPs and mCPs, respectively) NCLDVs), which are themselves parasites of unicellular eukaryotes. (12). In addition, genes encoding two different families of integrases This coupled parasitism can result in the indirect control of eukaryotic have been identified in several virophages: A putative rve integrase cell mortality by virophages. However, the details of such tripartite was found in Mavirus and ALM (8, 10), whereas Sputnik encodes a ∼ relationships remain largely unexplored. We have discovered 300 putative tyrosine integrase (1). Among virophage genes, only PRO, predicted genes of putative virophage origin in the nuclear genome ATPase, MCP, and mCP support the monophyly of virophages, of the unicellular alga Bigelowiella natans. Physical clustering of these whereas the remaining gene complement shows complex phyloge- genes indicates that virophage genomes are integrated into the B. natans genome. Virophage inserts show high levels of similarity nies suggestive of gene replacement (12). -

23.3 Groups of Protists
Chapter 23 | Protists 639 cysts that are a protective, resting stage. Depending on habitat of the species, the cysts may be particularly resistant to temperature extremes, desiccation, or low pH. This strategy allows certain protists to “wait out” stressors until their environment becomes more favorable for survival or until they are carried (such as by wind, water, or transport on a larger organism) to a different environment, because cysts exhibit virtually no cellular metabolism. Protist life cycles range from simple to extremely elaborate. Certain parasitic protists have complicated life cycles and must infect different host species at different developmental stages to complete their life cycle. Some protists are unicellular in the haploid form and multicellular in the diploid form, a strategy employed by animals. Other protists have multicellular stages in both haploid and diploid forms, a strategy called alternation of generations, analogous to that used by plants. Habitats Nearly all protists exist in some type of aquatic environment, including freshwater and marine environments, damp soil, and even snow. Several protist species are parasites that infect animals or plants. A few protist species live on dead organisms or their wastes, and contribute to their decay. 23.3 | Groups of Protists By the end of this section, you will be able to do the following: • Describe representative protist organisms from each of the six presently recognized supergroups of eukaryotes • Identify the evolutionary relationships of plants, animals, and fungi within the six presently recognized supergroups of eukaryotes • Identify defining features of protists in each of the six supergroups of eukaryotes. In the span of several decades, the Kingdom Protista has been disassembled because sequence analyses have revealed new genetic (and therefore evolutionary) relationships among these eukaryotes. -

New Phylogenomic Analysis of the Enigmatic Phylum Telonemia Further Resolves the Eukaryote Tree of Life
bioRxiv preprint doi: https://doi.org/10.1101/403329; this version posted August 30, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. New phylogenomic analysis of the enigmatic phylum Telonemia further resolves the eukaryote tree of life Jürgen F. H. Strassert1, Mahwash Jamy1, Alexander P. Mylnikov2, Denis V. Tikhonenkov2, Fabien Burki1,* 1Department of Organismal Biology, Program in Systematic Biology, Uppsala University, Uppsala, Sweden 2Institute for Biology of Inland Waters, Russian Academy of Sciences, Borok, Yaroslavl Region, Russia *Corresponding author: E-mail: [email protected] Keywords: TSAR, Telonemia, phylogenomics, eukaryotes, tree of life, protists bioRxiv preprint doi: https://doi.org/10.1101/403329; this version posted August 30, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Abstract The broad-scale tree of eukaryotes is constantly improving, but the evolutionary origin of several major groups remains unknown. Resolving the phylogenetic position of these ‘orphan’ groups is important, especially those that originated early in evolution, because they represent missing evolutionary links between established groups. Telonemia is one such orphan taxon for which little is known. The group is composed of molecularly diverse biflagellated protists, often prevalent although not abundant in aquatic environments. -

Gymnochlora Dimorpha Sp. Nov., a Chlorarachniophyte with Unique Daughter Cell Behaviour
Phycologia (2011) Volume 50 (3), 317–326 Published 3 May 2011 Gymnochlora dimorpha sp. nov., a chlorarachniophyte with unique daughter cell behaviour 1,2 3 1 SHUHEI OTA *{,ASTUKO KUDO AND KEN-ICHIRO ISHIDA 1Institute of Biological Sciences, Graduate School of Life and Environmental Sciences, University of Tsukuba, 1-1-1, Tennodai, Tsukuba 305-8572, Japan 2Station Biologique de Roscoff, Universite´ Pierre et Marie Curie (Paris 06), CNRS and UMR 7144, Place Georges Tessier, 29682 Roscoff, France 3The College of Biological Sciences, University of Tsukuba, 1-1-1, Tennodai, Tsukuba 305-8572, Japan OTA S., KUDO A. AND ISHIDA K.-I. 2011. Gymnochlora dimorpha sp. nov., a chlorarachniophyte with unique daughter cell behaviour. Phycologia 50: 317–326. DOI: 10.2216/09-102.1 A new chlorarachniophyte species, Gymnochlora dimorpha sp. nov., was described. This new species was isolated from an enrichment preculture of Padina sp. collected from a subtidal coral reef zone in Republic of Palau. The new strain, P314, was characterized by light and electron microscopy in the present study. Under the culture conditions used here, the amoeboid stage was dominant. Two types of amoeboid cells were found in the cultures: motile and flattened nonmotile (sessile) cells. The motile cells typically multiplied via binary cell division. The sessile cells were always present in the cultures, but they never became dominant under the culture conditions. Time-lapse video microscopic observations revealed that after cell division of a sessile cell, one daughter cell became motile, while the other remained sessile. According to ultrastructural characteristics of the pyrenoids and nucleomorphs, the new chlorarachniophyte strain belongs to the genus Gymnochlora. -

VII EUROPEAN CONGRESS of PROTISTOLOGY in Partnership with the INTERNATIONAL SOCIETY of PROTISTOLOGISTS (VII ECOP - ISOP Joint Meeting)
See discussions, stats, and author profiles for this publication at: https://www.researchgate.net/publication/283484592 FINAL PROGRAMME AND ABSTRACTS BOOK - VII EUROPEAN CONGRESS OF PROTISTOLOGY in partnership with THE INTERNATIONAL SOCIETY OF PROTISTOLOGISTS (VII ECOP - ISOP Joint Meeting) Conference Paper · September 2015 CITATIONS READS 0 620 1 author: Aurelio Serrano Institute of Plant Biochemistry and Photosynthesis, Joint Center CSIC-Univ. of Seville, Spain 157 PUBLICATIONS 1,824 CITATIONS SEE PROFILE Some of the authors of this publication are also working on these related projects: Use Tetrahymena as a model stress study View project Characterization of true-branching cyanobacteria from geothermal sites and hot springs of Costa Rica View project All content following this page was uploaded by Aurelio Serrano on 04 November 2015. The user has requested enhancement of the downloaded file. VII ECOP - ISOP Joint Meeting / 1 Content VII ECOP - ISOP Joint Meeting ORGANIZING COMMITTEES / 3 WELCOME ADDRESS / 4 CONGRESS USEFUL / 5 INFORMATION SOCIAL PROGRAMME / 12 CITY OF SEVILLE / 14 PROGRAMME OVERVIEW / 18 CONGRESS PROGRAMME / 19 Opening Ceremony / 19 Plenary Lectures / 19 Symposia and Workshops / 20 Special Sessions - Oral Presentations / 35 by PhD Students and Young Postdocts General Oral Sessions / 37 Poster Sessions / 42 ABSTRACTS / 57 Plenary Lectures / 57 Oral Presentations / 66 Posters / 231 AUTHOR INDEX / 423 ACKNOWLEDGMENTS-CREDITS / 429 President of the Organizing Committee Secretary of the Organizing Committee Dr. Aurelio Serrano -

Plastid-Targeting Peptides from the Chlorarachniophyte Bigelowiella Natans
J. Eukaryot. Microbiol., 51(5), 2004 pp. 529±535 q 2004 by the Society of Protozoologists Plastid-Targeting Peptides from the Chlorarachniophyte Bigelowiella natans MATTHEW B. ROGERS,a JOHN M. ARCHIBALD,a,1 MATTHEW A. FIELD,a CATHERINE LI,b BORIS STRIEPENb and PATRICK J. KEELINGa aCanadian Institute for Advanced Research, Department of Botany, University of British Columbia, 3529-6270 University Boulevard, Vancouver, BC, V6T 1Z4, Canada, and bCenter for Tropical & Emerging Global Diseases & Department of Cellular Biology, University of Georgia, 724 Biological Sciences Building, Athens, Georgia 30602, USA ABSTRACT. Chlorarachniophytes are marine amoebo¯agellate protists that have acquired their plastid (chloroplast) through secondary endosymbiosis with a green alga. Like other algae, most of the proteins necessary for plastid function are encoded in the nuclear genome of the secondary host. These proteins are targeted to the organelle using a bipartite leader sequence consisting of a signal peptide (allowing entry in to the endomembrane system) and a chloroplast transit peptide (for transport across the chloroplast envelope mem- branes). We have examined the leader sequences from 45 full-length predicted plastid-targeted proteins from the chlorarachniophyte Bigelowiella natans with the goal of understanding important features of these sequences and possible conserved motifs. The chemical characteristics of these sequences were compared with a set of 10 B. natans endomembrane-targeted proteins and 38 cytosolic or nuclear proteins, which show that the signal peptides are similar to those of most other eukaryotes, while the transit peptides differ from those of other algae in some characteristics. Consistent with this, the leader sequence from one B. -
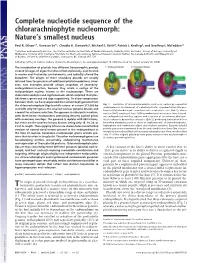
Complete Nucleotide Sequence of the Chlorarachniophyte Nucleomorph: Nature’S Smallest Nucleus
Complete nucleotide sequence of the chlorarachniophyte nucleomorph: Nature’s smallest nucleus Paul R. Gilson*†, Vanessa Su†‡, Claudio H. Slamovits§, Michael E. Reith¶, Patrick J. Keeling§, and Geoffrey I. McFadden‡ʈ *Infection and Immunity Division, The Walter and Eliza Hall Institute of Medical Research, Parkville 3050, Australia; ‡School of Botany, University of Melbourne, Victoria 3010, Australia; ¶Institute for Marine Biosciences, National Research Council, Halifax, NS, Canada B3H 3Z1; and §Department of Botany, University of British Columbia, Vancouver, BC, Canada V6T 1Z4 Edited by Jeffrey D. Palmer, Indiana University, Bloomington, IN, and approved April 19, 2006 (received for review January 26, 2006) The introduction of plastids into different heterotrophic protists created lineages of algae that diversified explosively, proliferated in marine and freshwater environments, and radically altered the biosphere. The origins of these secondary plastids are usually inferred from the presence of additional plastid membranes. How- ever, two examples provide unique snapshots of secondary- endosymbiosis-in-action, because they retain a vestige of the endosymbiont nucleus known as the nucleomorph. These are chlorarachniophytes and cryptomonads, which acquired their plas- tids from a green and red alga respectively. To allow comparisons between them, we have sequenced the nucleomorph genome from the chlorarachniophyte Bigelowiella natans: at a mere 373,000 bp Fig. 1. Evolution of chlorarachniophytes such as B. natans by sequential endosymbioses. Enslavement of a photosynthetic, cyanobacterium-like pro- and with only 331 genes, the smallest nuclear genome known and karyote (Cb) introduces photosynthesis into a eukaryotic host (Euk 1), whose a model for extreme reduction. The genome is eukaryotic in nature, nucleus (Nu1) acquires at least 1,000 cyanobacterial genes over time. -

Single Cell Transcriptomics, Mega-Phylogeny and the Genetic Basis Of
bioRxiv preprint doi: https://doi.org/10.1101/064030; this version posted July 15, 2016. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 Single cell transcriptomics, mega-phylogeny and the genetic basis of 2 morphological innovations in Rhizaria 3 4 Anders K. Krabberød1, Russell J. S. Orr1, Jon Bråte1, Tom Kristensen1, Kjell R. Bjørklund2 & Kamran 5 ShalChian-Tabrizi1* 6 7 1Department of BiosCienCes, Centre for Integrative MiCrobial Evolution and Centre for EpigenetiCs, 8 Development and Evolution, University of Oslo, Norway 9 2Natural History Museum, Department of ResearCh and ColleCtions University of Oslo, Norway 10 11 *Corresponding author: 12 Kamran ShalChian-Tabrizi 13 [email protected] 14 Mobile: + 47 41045328 15 16 17 Keywords: Cytoskeleton, phylogeny, protists, Radiolaria, Rhizaria, SAR, single-cell, transCriptomiCs 1 bioRxiv preprint doi: https://doi.org/10.1101/064030; this version posted July 15, 2016. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 18 Abstract 19 The innovation of the eukaryote Cytoskeleton enabled phagoCytosis, intracellular transport and 20 Cytokinesis, and is responsible for diverse eukaryotiC morphologies. Still, the relationship between 21 phenotypiC innovations in the Cytoskeleton and their underlying genotype is poorly understood. 22 To explore the genetiC meChanism of morphologiCal evolution of the eukaryotiC Cytoskeleton we 23 provide the first single Cell transCriptomes from unCultivable, free-living uniCellular eukaryotes: the 24 radiolarian speCies Lithomelissa setosa and Sticholonche zanclea. -

Living Organisms Author Their Read-Write Genomes in Evolution
biology Review Living Organisms Author Their Read-Write Genomes in Evolution James A. Shapiro ID Department of Biochemistry and Molecular Biology, University of Chicago GCIS W123B, 979 E. 57th Street, Chicago, IL 60637, USA; [email protected]; Tel.: +1-773-702-1625 Academic Editor: Andrés Moya Received: 23 August 2017; Accepted: 28 November 2017; Published: 6 December 2017 Abstract: Evolutionary variations generating phenotypic adaptations and novel taxa resulted from complex cellular activities altering genome content and expression: (i) Symbiogenetic cell mergers producing the mitochondrion-bearing ancestor of eukaryotes and chloroplast-bearing ancestors of photosynthetic eukaryotes; (ii) interspecific hybridizations and genome doublings generating new species and adaptive radiations of higher plants and animals; and, (iii) interspecific horizontal DNA transfer encoding virtually all of the cellular functions between organisms and their viruses in all domains of life. Consequently, assuming that evolutionary processes occur in isolated genomes of individual species has become an unrealistic abstraction. Adaptive variations also involved natural genetic engineering of mobile DNA elements to rewire regulatory networks. In the most highly evolved organisms, biological complexity scales with “non-coding” DNA content more closely than with protein-coding capacity. Coincidentally, we have learned how so-called “non-coding” RNAs that are rich in repetitive mobile DNA sequences are key regulators of complex phenotypes. Both biotic and abiotic ecological challenges serve as triggers for episodes of elevated genome change. The intersections of cell activities, biosphere interactions, horizontal DNA transfers, and non-random Read-Write genome modifications by natural genetic engineering provide a rich molecular and biological foundation for understanding how ecological disruptions can stimulate productive, often abrupt, evolutionary transformations.