Unsupervised and Supervised Principal Component Analysis: Tutorial
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Matrix Analysis and Applications
MATRIX ANALYSIS APPLICATIONS XIAN-DA ZHANG Tsinghua University, Beijing H Cambridge UNIVERSITY PRESS Contents Preface page xvii Notation xxi Abbreviations xxxi Algorithms xxxiv PART I MATRIX ALGEBRA 1 1 Introduction to Matrix Algebra 3 1.1 Basic Concepts of Vectors and Matrices 3 1.1.1 Vectors and Matrices 3 1.1.2 Basic Vector Calculus 6 1.1.3 Basic Matrix Calculus 8 1.1.4 Linear Independence of Vectors 11 1.1.5 Matrix Functions 11 1.2 Elementary Row Operations and Applications 13 1.2.1 Elementary Row Operations 13 1.2.2 Gauss Elimination Methods 16 1.3 Sets, Vector Subspaces and Linear Mapping 20 1.3.1 Sets 20 1.3.2 Fields and Vector Spaces 22 1.3.3 Linear Mapping 24 1.4 Inner Products and Vector Norms 27 1.4.1 Inner Products of Vectors 27 1.4.2 Norms of Vectors 28 1.4.3 Similarity Comparison Between Vectors 32 1.4.4 Banach Space, Euclidean Space, Hilbert Space 35 1.4.5 Inner Products and Norms of Matrices 36 1.5 Random Vectors 40 1.5.1 Statistical Interpretation of Random Vectors 41 1.5.2 Gaussian Random Vectors 44 viii 1.6 Performance Indexes of Matrices 47 1.6.1 Quadratic Forms 47 1.6.2 Determinants 49 1.6.3 Matrix Eigenvalues 52 1.6.4 Matrix Trace 54 1.6.5 Matrix Rank 56 1.7 Inverse Matrices and Pseudo-Inverse Matrices 59 1.7.1 Definition and Properties of Inverse Matrices 59 1.7.2 Matrix Inversion Lemma 60 1.7.3 Inversion of Hermitian Matrices 61 1.7.4 Left and Right Pseudo-Inverse Matrices 63 1.8 Moore-Penrose Inverse Matrices 65 1.8.1 Definition and Properties 65 1.8.2 Computation of Moore-Penrose Inverse Matrix 69 1.9 Direct Sum -

Lecture 0. Useful Matrix Theory
Lecture 0. Useful matrix theory Basic definitions A p × 1 vector a is a column vector of real (or complex) numbers a1; : : : ; ap, 0 1 a1 B . C a = @ . A ap By default, a vector is understood as a column vector. The collection of such column vectors is called the vector space of dimension p and is denoted by Rp (or Cp). An 1 × p row vector is obtained by the transpose of the column vector and is 0 a = (a1; : : : ; ap): A p × q matrix A is a rectangular array of real (or complex) numbers a11; a12; : : : ; apq written as 2 3 a11 ··· a1q 6 . 7 A = 4 . 5 = (aij)(p×q); ap1 ··· apq so that aij is the element in the ith row and jth column of A. The following definitions are basic: 1. A is a square matrix if p = q. 2. A = 0 is a zero matrix if aij = 0 for all i; j. 3. A = I = Ip is the identity matrix of order p if A is a square matrix of order p and aij = 0 for i 6= j and aij = 1 for i = j. 4. The transpose of a p × q matrix A, denoted by A0 or AT , is the q × p matrix by 0 0 0 0 interchanging the rows and columns of A, A = (aji). Note that (AB) = B A and (A0)0 = A. 5. A square matrix A is called symmetric if A = A0. 6. A square matrix A is called skew-symmetric if A = −A0. 7. A square matrix A is called diagonal if aij = 0 for all i 6= j and is denoted by diag(a11; : : : ; app). -
Notes on Matrix Algebra
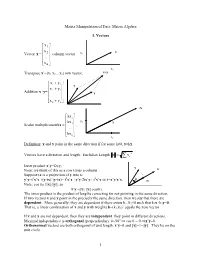
Matrix Manipulation of Data: Matrix Algebra 1. Vectors ⎡ x1 ⎤ ⎢ ⎥ x 2 x Vector x = ⎢ ⎥ column vector x2 ⎢ .. ⎥ ⎢ ⎥ ⎣x n ⎦ x1 x+y Transpose x’=[x1 x2…xn] row vector, ⎡ x+ y ⎤ 1 1 y ⎢x+ y ⎥ Addition x+y= ⎢ 2 2 ⎥ ⎢ .. ⎥ x ⎢ ⎥ ⎣xn+ y n ⎦ 2x ⎡kx1 ⎤ ⎢ ⎥ kx x2 x Scalar multiplication kx = ⎢ 2 ⎥ , ⎢ .. ⎥ ⎢ ⎥ ⎣kx n ⎦ x 1 Definition: x and y point in the same direction if for some k≠0, y=kx. 2 Vectors have a direction and length. Euclidian Length x= ∑ x i Inner product x’y=Σx y i i y Note: we think of this as a row times a column x Suppose tx is a projection of y into x: 2 2 2 y’y=t x’x +(y-tx)’(y-tx)= t x’x +y’y-2tx’y+ t x’x ⇒ t=x’y/x’x. θ tx Note: cos θ≡ t||x||/||y||, so x’y =||x|| ||y|| cos(θ). The inner product is the product of lengths correcting for not pointing in the same direction. If two vectors x and y point in the precisely the same direction, then we say that there are dependent. More generally, they are dependent if there exists k1, k2≠0 such that k1x+k2y=0. That is, a linear combination of x and y with weights k=(k1,k2)’ equals the zero vector If x and y are not dependent, then they are independent: they point in different directions. Maximal independence is orthogonal (perpendicular): θ=90o ⇒ cos θ = 0 ⇒x’y=0. Orthonormal vectors are both orthogonal of unit length: x’y=0 and ||x||=1=||y||. -

Iso-Charts: Stretch-Driven Mesh Parameterization Using Spectral Analysis
Eurographics Symposium on Geometry Processing (2004) R. Scopigno, D. Zorin, (Editors) Iso-charts: Stretch-driven Mesh Parameterization using Spectral Analysis Kun Zhou 1 John Synder 2 Baining Guo 1 Heung-Yeung Shum 1 1 Microsoft Research Asia 2 Microsoft Research Abstract We describe a fully automatic method, called iso-charts, to create texture atlases on arbitrary meshes. It is the first to consider stretch not only when parameterizing charts, but also when forming charts. The output atlas bounds stretch by a user-specified constant, allowing the user to balance the number of charts against their stretch. Our approach combines two seemingly incompatible techniques: stretch-minimizing parameterization, based on the surface integral of the trace of the local metric tensor, and the “isomap” or MDS (multi-dimensional scaling) parameterization, based on an eigen-analysis of the matrix of squared geodesic distances between pairs of mesh vertices. We show that only a few iterations of nonlinear stretch optimization need be applied to the MDS param- eterization to obtain low-stretch atlases. The close relationship we discover between these two parameterizations also allows us to apply spectral clustering based on MDS to partition the mesh into charts having low stretch. We also novelly apply the graph cut algorithm in optimizing chart boundaries to further minimize stretch, follow sharp features, and avoid meandering. Overall, our algorithm creates texture atlases quickly, with fewer charts and lower stretch than previous methods, providing improvement in applications like geometric remeshing. We also describe an extension, signal-specialized atlas creation, to efficiently sample surface signals, and show for the first time that considering signal stretch in chart formation produces better texture maps. -

Simple Matrices
Appendix B Simple matrices Mathematicians also attempted to develop algebra of vectors but there was no natural definition of the product of two vectors that held in arbitrary dimensions. The first vector algebra that involved a noncommutative vector product (that is, v w need not equal w v) was proposed by Hermann Grassmann in his book× Ausdehnungslehre× (1844 ). Grassmann’s text also introduced the product of a column matrix and a row matrix, which resulted in what is now called a simple or a rank-one matrix. In the late 19th century the American mathematical physicist Willard Gibbs published his famous treatise on vector analysis. In that treatise Gibbs represented general matrices, which he called dyadics, as sums of simple matrices, which Gibbs called dyads. Later the physicist P. A. M. Dirac introduced the term “bra-ket” for what we now call the scalar product of a “bra” (row) vector times a “ket” (column) vector and the term “ket-bra” for the product of a ket times a bra, resulting in what we now call a simple matrix, as above. Our convention of identifying column matrices and vectors was introduced by physicists in the 20th century. Suddhendu Biswas [52, p.2] − B.1 Rank-1 matrix (dyad) Any matrix formed from the unsigned outer product of two vectors, T M N Ψ = uv R × (1783) ∈ where u RM and v RN , is rank-1 and called dyad. Conversely, any rank-1 matrix must have the∈ form Ψ . [233∈, prob.1.4.1] Product uvT is a negative dyad. For matrix products ABT, in general, we have − (ABT) (A) , (ABT) (BT) (1784) R ⊆ R N ⊇ N B.1 § with equality when B =A [374, §3.3, 3.6] or respectively when B is invertible and (A)= 0. -

APPENDIX a Matrix Algebra
Greene-2140242 book December 1, 2010 8:8 APPENDIXQ A MATRIX ALGEBRA A.1 TERMINOLOGY A matrix is a rectangular array of numbers, denoted ⎡ ⎤ a11 a12 ··· a1K ⎢ ··· ⎥ = = = ⎣a21 a22 a2K ⎦ . A [aik] [A]ik ··· (A-1) an1 an2 ··· anK The typical element is used to denote the matrix. A subscripted element of a matrix is always read as arow,column. An example is given in Table A.1. In these data, the rows are identified with years and the columns with particular variables. A vector is an ordered set of numbers arranged either in a row or a column. In view of the preceding, a row vector is also a matrix with one row, whereas a column vector is a matrix with one column. Thus, in Table A.1, the five variables observed for 1972 (including the date) constitute a row vector, whereas the time series of nine values for consumption is a column vector. A matrix can also be viewed as a set of column vectors or as a set of row vectors.1 The dimensions of a matrix are the numbers of rows and columns it contains. “A is an n × K matrix” (read “n by K”) will always mean that A has n rows and K columns. If n equals K, then A is a square matrix. Several particular types of square matrices occur frequently in econometrics. • A symmetric matrix is one in which aik = aki for all i and k. • A diagonal matrix is a square matrix whose only nonzero elements appear on the main diagonal, that is, moving from upper left to lower right. -

Data Sample Matrices
Data Sample Matrices Professor Dan A. Simovici UMB Professor Dan A. Simovici (UMB) Data Sample Matrices 1 / 29 1 The Sample Matrix 2 The Covariance Matrix Professor Dan A. Simovici (UMB) Data Sample Matrices 2 / 29 Matrices as organizers for data sets n Data set: a sequence E of m vectors of R , (u1,...,um). th The j components (u i )j of these vectors correspond to the values of a random variable Vj , where 1 ≤ j ≤ n. This data series will be represented as a sample matrix having m rows 0 0 u1,...,um and n columns v 1,...,v n. The number m is the size of the sample. Professor Dan A. Simovici (UMB) Data Sample Matrices 3 / 29 Rows, Experiments, Attributes 0 Each row vector ui corresponds to an experiment Ei in the series of experiments E = (E1,..., Em); the experiment Ei consists of measuring 0 the n components of ui = (xi1,..., xin): v 1 · · · v n 0 u1 x11 · · · x1n 0 u2 x21 · · · x2n . 0 um xm1 · · · xmn The column vector x1j x2j v j = . . xmj th represents the measurements of the j variable Vj of the experiment, for 1 ≤ j ≤ n, as shown below. These variables are usually referred to as attributes or features of the series E. Professor Dan A. Simovici (UMB) Data Sample Matrices 4 / 29 Definition The sample matrix of E is the matrix X ∈ Cm×n given by 0 u1 . X = . = (v 1 · · · v n). 0 um Professor Dan A. Simovici (UMB) Data Sample Matrices 5 / 29 Pairwise Distances Pairwise distances between the row vectors of X ∈ Rm×n can be computed with the MATLAB function pdist(X) . -

Matrix Analysis and Applications Xian-Da Zhang Frontmatter More Information
Cambridge University Press 978-1-108-41741-9 — Matrix Analysis and Applications Xian-Da Zhang Frontmatter More Information MATRIX ANALYSIS AND APPLICATIONS This balanced and comprehensive study presents the theory, methods and applications of matrix analysis in a new theoretical framework, allowing readers to understand second- order and higher-order matrix analysis in a completely new light. Alongside the core subjects in matrix analysis, such as singular value analysis, the solu- tion of matrix equations and eigenanalysis, the author introduces new applications and perspectives that are unique to this book. The very topical subjects of gradient analysis and optimization play a central role here. Also included are subspace analysis, projection analysis and tensor analysis, subjects which are often neglected in other books. Having provided a solid foundation to the subject, the author goes on to place particular emphasis on the many applications matrix analysis has in science and engineering, making this book suitable for scientists, engineers and graduate students alike. XIAN- DA ZHANG is Professor Emeritus in the Department of Automation, at Tsinghua University, Beijing. He was a Distinguished Professor at Xidian University, Xi’an, China – a post awarded by the Ministry of Education of China, and funded by the Ministry of Education of China and the Cheung Kong Scholars Programme – from 1999 to 2002. His areas of research include signal processing, pattern recognition, machine learning and related applied mathematics. He has published over 120 international journal and confer- ence papers, and 7 books in Chinese. He taught the graduate course “Matrix Analysis and Applications” at Tsinghua University from 2004 to 2011. -
Linear Algebra Notes
Stat 159/259: Linear Algebra Notes Jarrod Millman November 16, 2015 Abstract These notes assume you’ve taken a semester of undergraduate linear algebra. In particular, I assume you are familiar with the following: • solving systems of linear equations using Gaussian elimination • linear combinations of vectors to produce a space • the rank of a matrix • the Gram–Schmidt process for orthonormalising a set of vectors • eigenvectors and eigenvalues of a square matrix I will briefly review some of the above topics, but if you haven’t seen them before the presentation may be difficult to follow. If so, please review your introductory linear algebra textbook or use Wikipedia. Background Introductory linear algebra texts often start with an examination of simultaneous systems of linear equations. For example, consider the following system of linear equations 2x1 + x2 = 3 x1 − 3x2 = −2. For convenience, we abbreviate this system 2 1 x1 3 = 1 −3 x2 −2 | {z } | {z } | {z } A x b where A is the coefficient matrix, x is a column vector of unknowns, and b is a column vector of constants. 1 You should verify that 1 x = 1 solves the given system. More generally, a real-valued matrix A is an ordered, m × n rectangular array of real numbers, which when multiplied on the right by an n-dimensional column vector x produces an m-dimensional column vector b. Having introduced the Ax = b notation, we can view the m × n matrix A as a linear function from Rn to Rm. A linear function is one that respects proportions and for which the effect of a sum is the sum of individual effects. -

BU-1355-M.Pdf
MATRIX ALGEBRA S. R. Searle Biometrics Unit, Cornell University, Ithaca, N.Y. 14853 BU-1355-M July 1996 Abstract This is a 6,000-10,000 word article invited for an upcoming Encyclopedia of Biostatistics. It attempts to describe and summarize some of the main features of matrix algebra as are customarily found in the statistical literature. Key Words Basic matrix operations, special matrices, determinants, inverses, rank, canonical forms, generalized inverses, linear equations, eigenroots, eigenvectors, differential calculus, vee, vech, real and complex matrices, statistical uses. 1 MATRIX ALGEBRA The algebra we learn when teenagers has letters of the alphabet each representing a number. For example: a father and son are x andy years old, respectively, and their total age is 70. In ten year's time the father will be twice as old as the son. Hence x + y = 70 and x + 10 = 2(y + 10) and so x = 50 and y = 20. In contrast, matrix algebra is the algebra of letters each representing many numbers, with those numbers always arrayed in the form of a rectangle (or square). An example is 9 0 7 X- [ - u2 + v -3 6.1 GENERAL DESCRIPTION A matrix is a rectangular array of numbers, which can be any mixture of numbers that are complex, real, zero, positive, negative, decimal, fractions, or algebraic expressions. When none of them is complex (i.e., involving R), the matrix is said to be real. And because statistics deals with data, which are real numbers (especially biological data), almost all of this article applies to real matrices. Each number in a matrix is called an element: in being some representation of a single number it is called a scalar, to contrast with matrix which represents many numbers. -

A Least Squares Formulation for a Class of Generalized Eigenvalue Problems in Machine Learning
A Least Squares Formulation for a Class of Generalized Eigenvalue Problems in Machine Learning Liang Sun [email protected] Shuiwang Ji [email protected] Jieping Ye [email protected] Department of Computer Science and Engineering, Arizona State University, Tempe, AZ 85287, USA Abstract nant Analysis (LDA), and Hypergraph Spectral Learn- ing (HSL) (Hotelling, 1936; Rosipal & Kr¨amer, 2006; Many machine learning algorithms can be Sun et al., 2008a; Tao et al., 2009; Ye, 2007). Al- formulated as a generalized eigenvalue prob- though well-established algorithms in numerical linear lem. One major limitation of such formula- algebra have been developed to solve generalized eigen- tion is that the generalized eigenvalue prob- value problems, they are in general computationally lem is computationally expensive to solve expensive and hence may not scale to large-scale ma- especially for large-scale problems. In this chine learning problems. In addition, it is challenging paper, we show that under a mild condi- to directly incorporate the sparsity constraint into the tion, a class of generalized eigenvalue prob- mathematical formulation of these techniques. Spar- lems in machine learning can be formulated sity often leads to easy interpretation and a good gen- as a least squares problem. This class of eralization ability. It has been used successfully in lin- problems include classical techniques such as ear regression (Tibshirani, 1996), and Principal Com- Canonical Correlation Analysis (CCA), Par- ponent Analysis (PCA) (d’Aspremont et al., 2004). tial Least Squares (PLS), and Linear Dis- criminant Analysis (LDA), as well as Hy- Multivariate Linear Regression (MLR) that minimizes pergraph Spectral Learning (HSL). -
LARGEST CENTER-SPECIFIC MARGIN for DIMENSION REDUCTION Jian'an Zhang Yuan Yuan Feiping Nie Qi Wang School of Computer Science
LARGEST CENTER-SPECIFIC MARGIN FOR DIMENSION REDUCTION Jian’an Zhang Yuan Yuan Feiping Nie Qi Wang School of Computer Science and Center for OPTical IMagery Analysis and Learning (OPTIMAL), Northwestern Polytechnical University, Xi’an 710072, Shaanxi, PR China ABSTRACT 6DPSOHVIURPLWKFODVV Dimensionality reduction plays an important role in solving 6DPSOHVIURPMWKFODVV the “curse of the dimensionality” and attracts a number of 5HODWLRQVKLSVEHWZHHQ FHQWHUVDQGVDPSOHV researchers in the past decades. In this paper, we proposed 5HODWLRQVKLSVEHWZHHQ a new supervised linear dimensionality reduction method FHQWHUV named largest center-specific margin (LCM) based on the in- tuition that after linear transformation, the distances between Fig. 1. Diagram for the intuition, circles and triangles should the points and their corresponding class centers should be be close to their diamond centers and the distances between small enough, and at the same time the distances between d- different diamond centers should be as large as possible. ifferent unknown class centers should be as large as possible. On the basis of this observation, we take the unknown class centers into consideration for the first time and construct an optimization function to formulate this problem. In addition, we creatively transform the optimization objective function methods do not utilize the information of class centers. There- into a matrix function and solve the problem analytically. fore, supervised method with class centers’ information can Finally, experiment results on three real datasets show the be taken into consideration and applied into dimensionality competitive performance of our algorithm. reduction. Index Terms— Dimensionality Reduction, LCM, Center- In this work, we proposed a new linear dimensionality re- specific Method duction method on the basis of the observation that after lin- ear transformation the distances between the points and their corresponding class center should be small enough, and at 1.