A Comparison of Three Methods of Estimation Applied to Contaminated Circular Data
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Transformation of Circular Random Variables Based on Circular Distribution Functions
Filomat 32:17 (2018), 5931–5947 Published by Faculty of Sciences and Mathematics, https://doi.org/10.2298/FIL1817931M University of Nis,ˇ Serbia Available at: http://www.pmf.ni.ac.rs/filomat Transformation of Circular Random Variables Based on Circular Distribution Functions Hatami Mojtabaa, Alamatsaz Mohammad Hosseina a Department of Statistics, University of Isfahan, Isfahan, 8174673441, Iran Abstract. In this paper, we propose a new transformation of circular random variables based on circular distribution functions, which we shall call inverse distribution function (id f ) transformation. We show that Mobius¨ transformation is a special case of our id f transformation. Very general results are provided for the properties of the proposed family of id f transformations, including their trigonometric moments, maximum entropy, random variate generation, finite mixture and modality properties. In particular, we shall focus our attention on a subfamily of the general family when id f transformation is based on the cardioid circular distribution function. Modality and shape properties are investigated for this subfamily. In addition, we obtain further statistical properties for the resulting distribution by applying the id f transformation to a random variable following a von Mises distribution. In fact, we shall introduce the Cardioid-von Mises (CvM) distribution and estimate its parameters by the maximum likelihood method. Finally, an application of CvM family and its inferential methods are illustrated using a real data set containing times of gun crimes in Pittsburgh, Pennsylvania. 1. Introduction There are various general methods that can be used to produce circular distributions. One popular way is based on families defined on the real line, i.e., linear distributions such as normal, Cauchy and etc. -

211365757.Pdf
; i LJ .u June 1967 LJ NONPARAMETRIC TESTS OF LOCATION 1 FOR CIRCULAR DISTRIBUTIONS LJ Siegfried Schach , I 6J Technical Report No. 95 ui I u ui i u u University of Minnesota u Minneapolis, Minnesota I I I I ~ :w I 1 Research supported by the National Science Foundation under Grant No. GP-3816 u and GP-6859, and U.S. Navy Grant NONR(G)-00003-66. I I LJ iw I ABSTRACT In this thesis classes of nonparametric tests for circular distributions are investigated. In the one-sample problem the null hypothesis states that a distribution is symmetric around the horizontal axis. An interesting class of alternatives consists of shifts of such distributions by a certain angle ~ * O. In the two-sample case the null hypothesis claims that two samples have originated from the same underlying distributiono The alternatives considered consist of pairs of underlying distributions such that one of them is obtained from the other by a shift of the probability mass by an angle ~ * O. The general outline of the reasoning is about the same for the two cases: We first define (Sections 2 and 8) a suitable group of transfor mations of the sample space, which is large enough to make any invariant test nonparametric, in the sense that any invariant test statistic has the same distribution for all elements of the null hypothesiso We then derive, for parametric classes of distributions, the efficacy of the best parametric test, in order to have a standard of comparison for nonparametric tests (Sections 3 and 9). Next we find a locally most powerful invariant test against shift alternatives (Sectiora4 and 10). -

Wrapped Weighted Exponential Distribution
Int. Statistical Inst.: Proc. 58th World Statistical Congress, 2011, Dublin (Session CPS046) p.5019 Wrapped weighted exponential distribution Roy, Shongkour Department of Statistics, Jahangirnagar University 309, Bangabandhu Hall, Dhaka 1342, Bangladesh. E- mail: [email protected] Adnan, Mian Department of Statistics, Jahangirnagar University Dhaka 1342, Bangladesh. 1. Introduction Weighted exponential distribution was first derived by R. D. Gupta and D. Kundu (2009). It has extensive uses in Biostatistics, and life science, etc. Since maximum important variables are axial form in bird migration study, the study of weighted exponential distribution in case of circular data can be a better perspective. Although wrapped distribution was first initiated by (Levy P.L., 1939), a few number of authors (Mardia, 972), (Rao Jammalmadaka and SenGupta, 2001), (Cohello, 2007), (Shongkour and Adnan, 2010, 2011) worked on wrapped distributions. Their works include various wrapped distributions such as wrapped exponential, wrapped gamma, wrapped chi- square, etc. However, wrapped weighted exponential distribution has never been premeditated before, although this distribution may ever be a better one to model bird orientation data. 2. Definition and derivation A random variable X (in real line) has weighted exponential distributions if its probability density functions of form α +1 fx()=−−>>>λα e−λx () 1 exp()λαλx ; x 0, 0, 0 α where α is shape parameter and λ is scale parameter. Let us consider, θ ≡=θπ()xx (mod2) (2.1) then θ is wrapped (around the circle) or wrapped weighted exponential circular random variables that for θ ∈[0,2π ) has probability density function ∞ gfm()θ =+∑ X (θπ 2 ) (2.2) m=0 α +1 −−λθ∞ 2m πλ −+αλ() θ2m π =−λ ee∑ (1 e ) ; 02,0,0≤ θ ≤>>πα λ α m=0 where the random variable θ has a wrapped weighted exponential distribution with parameter and the above pdf. -

Recent Advances in Directional Statistics
Recent advances in directional statistics Arthur Pewsey1;3 and Eduardo García-Portugués2 Abstract Mainstream statistical methodology is generally applicable to data observed in Euclidean space. There are, however, numerous contexts of considerable scientific interest in which the natural supports for the data under consideration are Riemannian manifolds like the unit circle, torus, sphere and their extensions. Typically, such data can be represented using one or more directions, and directional statistics is the branch of statistics that deals with their analysis. In this paper we provide a review of the many recent developments in the field since the publication of Mardia and Jupp (1999), still the most comprehensive text on directional statistics. Many of those developments have been stimulated by interesting applications in fields as diverse as astronomy, medicine, genetics, neurology, aeronautics, acoustics, image analysis, text mining, environmetrics, and machine learning. We begin by considering developments for the exploratory analysis of directional data before progressing to distributional models, general approaches to inference, hypothesis testing, regression, nonparametric curve estimation, methods for dimension reduction, classification and clustering, and the modelling of time series, spatial and spatio- temporal data. An overview of currently available software for analysing directional data is also provided, and potential future developments discussed. Keywords: Classification; Clustering; Dimension reduction; Distributional -

Chapter 2 Parameter Estimation for Wrapped Distributions
ABSTRACT RAVINDRAN, PALANIKUMAR. Bayesian Analysis of Circular Data Using Wrapped Dis- tributions. (Under the direction of Associate Professor Sujit K. Ghosh). Circular data arise in a number of different areas such as geological, meteorolog- ical, biological and industrial sciences. We cannot use standard statistical techniques to model circular data, due to the circular geometry of the sample space. One of the com- mon methods used to analyze such data is the wrapping approach. Using the wrapping approach, we assume that, by wrapping a probability distribution from the real line onto the circle, we obtain the probability distribution for circular data. This approach creates a vast class of probability distributions that are flexible to account for different features of circular data. However, the likelihood-based inference for such distributions can be very complicated and computationally intensive. The EM algorithm used to compute the MLE is feasible, but is computationally unsatisfactory. Instead, we use Markov Chain Monte Carlo (MCMC) methods with a data augmentation step, to overcome such computational difficul- ties. Given a probability distribution on the circle, we assume that the original distribution was distributed on the real line, and then wrapped onto the circle. If we can unwrap the distribution off the circle and obtain a distribution on the real line, then the standard sta- tistical techniques for data on the real line can be used. Our proposed methods are flexible and computationally efficient to fit a wide class of wrapped distributions. Furthermore, we can easily compute the usual summary statistics. We present extensive simulation studies to validate the performance of our method. -

A Half-Circular Distribution on a Circle (Taburan Separa-Bulat Dalam Bulatan)
Sains Malaysiana 48(4)(2019): 887–892 http://dx.doi.org/10.17576/jsm-2019-4804-21 A Half-Circular Distribution on a Circle (Taburan Separa-Bulat dalam Bulatan) ADZHAR RAMBLI*, IBRAHIM MOHAMED, KUNIO SHIMIZU & NORLINA MOHD RAMLI ABSTRACT Up to now, circular distributions are defined in [0,2 π), except for axial distributions on a semicircle. However, some circular data lie within just half of this range and thus may be better fitted by a half-circular distribution, which we propose and develop in this paper using the inverse stereographic projection technique on a gamma distributed variable. The basic properties of the distribution are derived while its parameters are estimated using the maximum likelihood estimation method. We show the practical value of the distribution by applying it to an eye data set obtained from a glaucoma clinic at the University of Malaya Medical Centre, Malaysia. Keywords: Gamma distribution; inverse stereographic projection; maximum likelihood estimation; trigonometric moments; unimodality ABSTRAK Sehingga kini, taburan bulatan ditakrifkan dalam [0,2 π), kecuali untuk taburan paksi aksial pada semi-bulatan. Walau bagaimanapun, terdapat data bulatan berada hanya separuh daripada julat ini dan ia lebih sesuai dengan taburan separuh-bulatan, maka kami mencadang dan membangunkan dalam kertas ini menggunakan teknik unjuran stereografik songsang pada pemboleh ubah taburan gamma. Sifat asas taburan diperoleh manakala parameter dinilai menggunakan kaedah anggaran kebolehjadian maksimum. Nilai praktikal taburan ini dipraktiskan pada set data mata yang diperoleh daripada klinik glaukoma di Pusat Kesihatan, Universiti Malaya, Malaysia. Kata kunci: Anggaran kebolehjadian maksimum; momen trigonometrik; taburan Gamma; unimodaliti; unjuran stereografik songsang INTRODUCTION symmetric unimodal distributions on the unit circle that Circular data refer to observations measured in radians or contains the uniform, von Mises, cardioid and wrapped degrees. -

A General Approach for Obtaining Wrapped Circular Distributions Via Mixtures
UC Santa Barbara UC Santa Barbara Previously Published Works Title A General Approach for obtaining wrapped circular distributions via mixtures Permalink https://escholarship.org/uc/item/5r7521z5 Journal Sankhya, 79 Author Jammalamadaka, Sreenivasa Rao Publication Date 2017 Peer reviewed eScholarship.org Powered by the California Digital Library University of California 1 23 Your article is protected by copyright and all rights are held exclusively by Indian Statistical Institute. This e-offprint is for personal use only and shall not be self- archived in electronic repositories. If you wish to self-archive your article, please use the accepted manuscript version for posting on your own website. You may further deposit the accepted manuscript version in any repository, provided it is only made publicly available 12 months after official publication or later and provided acknowledgement is given to the original source of publication and a link is inserted to the published article on Springer's website. The link must be accompanied by the following text: "The final publication is available at link.springer.com”. 1 23 Author's personal copy Sankhy¯a:TheIndianJournalofStatistics 2017, Volume 79-A, Part 1, pp. 133-157 c 2017, Indian Statistical Institute ! AGeneralApproachforObtainingWrappedCircular Distributions via Mixtures S. Rao Jammalamadaka University of California, Santa Barbara, USA Tomasz J. Kozubowski University of Nevada, Reno, USA Abstract We show that the operations of mixing and wrapping linear distributions around a unit circle commute, and can produce a wide variety of circular models. In particular, we show that many wrapped circular models studied in the literature can be obtained as scale mixtures of just the wrapped Gaussian and the wrapped exponential distributions, and inherit many properties from these two basic models. -

On Sobolev Tests of Uniformity on the Circle with an Extension to the Sphere
UC Santa Barbara UC Santa Barbara Previously Published Works Title On Sobolev tests of uniformity on the circle with an extension to the sphere Permalink https://escholarship.org/uc/item/7mb6w6k6 Journal BERNOULLI, 26(3) ISSN 1350-7265 Authors Jammalamadaka, Sreenivasa Rao Meintanis, Simos Verdebout, Thomas Publication Date 2020-08-01 DOI 10.3150/19-BEJ1191 Peer reviewed eScholarship.org Powered by the California Digital Library University of California Bernoulli 26(3), 2020, 2226–2252 https://doi.org/10.3150/19-BEJ1191 On Sobolev tests of uniformity on the circle with an extension to the sphere SREENIVASA RAO JAMMALAMADAKA1, SIMOS MEINTANIS2 and THOMAS VERDEBOUT3 1Department of Statistics and Applied Probability, University of California Santa Barbara, Santa Barbara, CA 93106-3110, USA. E-mail: [email protected] 2Department of Economics, National and Kapodistrian University of Athens,1, Sofokleous and Aristidou street, 10559 Athens, Greece, and Unit for Business Mathematics and Informatics, North-West University, Vaal Triangle Campus, PO Box 1174, Vanderbijlpark 1900, South Africa. E-mail: [email protected] 3ECARES and Département de Mathématique, Université libre de Bruxelles, Avenue F.D. Roosevelt, 50, ECARES, CP114/04, B-1050, Brussels, Belgium. E-mail: [email protected] Circular and spherical data arise in many applications, especially in biology, Earth sciences and astronomy. In dealing with such data, one of the preliminary steps before any further inference, is to test if such data is isotropic, that is, uniformly distributed around the circle or the sphere. In view of its importance, there is a considerable literature on the topic. In the present work, we provide new tests of uniformity on the circle based on original asymptotic results. -

Inferences Based on a Bivariate Distribution with Von Mises Marginals
Ann. Inst. Statist. Math. Vol. 57, No. 4, 789-802 (2005) (~)2005 The Institute of Statistical Mathematics INFERENCES BASED ON A BIVARIATE DISTRIBUTION WITH VON MISES MARGINALS GRACE S. SHIEH I AND RICHARD A. JOHNSON2 l Institute of Statistical Science, Academia Sinica, Taipei 115, Taiwan, R. O. C., e-mail: gshieh@stat .sinica.edu.tw 2Department of Statistics, University of Wisconsin-Madison, WI 53706, U.S.A., e-mail: [email protected] (Received April 21, 2004; revised August 24, 2004) Abstract. There is very little literature concerning modeling the correlation be- tween paired angular observations. We propose a bivariate model with von Mises marginal distributions. An algorithm for generating bivariate angles from this von Mises distribution is given. Maximum likelihood estimation is then addressed. We also develop a likelihood ratio test for independence in paired circular data. Applica- tion of the procedures to paired wind directions is illustrated. Employing simulation, using the proposed model, we compare the power of the likelihood ratio test with six existing tests of independence. Key words and phrases: Angular observations, maximum likelihood estimation, models of dependence, power, testing. 1. Introduction Testing for independence in paired circular (bivariate angular) data has been ex- plored by many statisticians; see Fisher (1995) and Mardia and Jupp (2000) for thorough accounts. Paired circular data are either both angular or both cyclic in nature. For in- stance, wind directions at 6 a.m. and at noon recorded at a weather station for 21 consecutive days (Johnson and Wehrly (1977)), estimated peak times for two successive measurements of diastolic blood pressure (Downs (1974)), among others. -

Comparing Measures of Fit for Circular Distributions
Comparing measures of fit for circular distributions by Zheng Sun B.Sc., Simon Fraser University, 2006 A Thesis Submitted in Partial Fulfillment of the Requirements for the Degree of MASTER OF SCIENCE in the Department of Mathematics and Statistics © Zheng Sun, 2009 University of Victoria All rights reserved. This dissertation may not be reproduced in whole or in part, by photocopying or other means, without the permission of the author. ii Comparing measures of fit for circular distributions by Zheng Sun B.Sc., Simon Fraser University, 2006 Supervisory Committee Dr. William J. Reed, Supervisor (Department of Mathematics and Statistics) Dr. Laura Cowen, Departmental Member (Department of Mathematics and Statistics) Dr. Mary Lesperance, Departmental Member (Department of Mathematics and Statistics) iii Supervisory Committee Dr. William J. Reed, Supervisor (Department of Mathematics and Statistics) Dr. Laura Cowen, Departmental Member (Department of Mathematics and Statistics) Dr. Mary Lesperance, Departmental Member (Department of Mathematics and Statistics) ABSTRACT This thesis shows how to test the fit of a data set to a number of different models, using Watson's U 2 statistic for both grouped and continuous data. While Watson's U 2 statistic was introduced for continuous data, in recent work, the statistic has been adapted for grouped data. However, when using Watson's U 2 for continuous data, the asymptotic distribution is difficult to obtain, particularly, for some skewed circular distributions that contain four or five parameters. Until now, U 2 asymptotic points are worked out only for uniform distribution and the von Mises distribution among all circular distributions. We give U 2 asymptotic points for the wrapped exponential distributions, and we show that U 2 asymptotic points when data are grouped is usually easier to obtain for other more advanced circular distributions. -
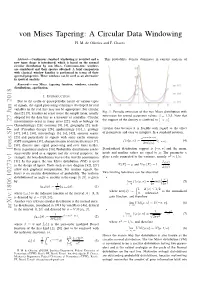
Von Mises Tapering: a Circular Data Windowing H
von Mises Tapering: A Circular Data Windowing H. M. de Oliveira and F. Chaves Abstract— Continuous standard windowing is revisited and a This probability density dominates in current analysis of new taper shape is introduced, which is based on the normal circular distribution by von Mises. Continuous-time windows are considered and their spectra obtained. A brief comparison with classical window families is performed in terms of their spectral properties. These windows can be used as an alternative in spectral analysis. Keywords— von Mises, tapering function, windows, circular distributions, apodization. I. INTRODUCTION Due to the cyclic or quasi-periodic nature of various types of signals, the signal processing techniques developed for real variables in the real line may not be appropriate. For circular data [2], [9], it makes no sense to use the sample mean, usually Fig. 1: Periodic extension of the von Mises distribution with β = adopted for the data line as a measure of centrality. Circular zero-mean for several parameter values: 1,3,5. Note that [−π; π] measurements occur in many areas [22], such as biology (or the support of the density is confined to . Chronobiology) [26], economy [8], [4], geography [5], med- ical (Circadian therapy [28], epidemiology [13]...), geology circular data because it is flexible with regard to the effect [47], [41], [34], meteorology [3], [6], [42], acoustic scatter of parameters and easy to interpret. In a standard notation, [23] and particularly in signals with some cyclic structure eκ cos(x−µ) (GPS navigation [31], characterization of oriented textures [7], f(xjµ, κ) := I[−π,π]: (4) 2πI0(κ) [38], discrete-time signal processing and over finite fields). -

On Some Circular Distributions Induced by Inverse Stereographic Projection
On Some Circular Distributions Induced by Inverse Stereographic Projection Shamal Chandra Karmaker A Thesis for The Department of Mathematics and Statistics Presented in Partial Fulfillment of the Requirements for the Degree of Master of Science (Mathematics) at Concordia University Montr´eal, Qu´ebec, Canada November 2016 c Shamal Chandra Karmaker 2016 CONCORDIA UNIVERSITY School of Graduate Studies This is to certify that the thesis prepared By: Shamal Chandra Karmaker Entitled: On Some Circular Distributions Induced by Inverse Stereographic Projection and submitted in partial fulfillment of the requirements for the degree of Master of Science (Mathematics) complies with the regulations of the University and meets the accepted standards with respect to originality and quality. Signed by the final examining committee: Examiner Dr. Arusharka Sen Examiner Dr. Wei Sun Thesis Supervisor Dr. Y.P. Chaubey Thesis Co-supervisor Dr. L. Kakinami Approved by Chair of Department or Graduate Program Director Dean of Faculty Date Abstract On Some Circular Distributions Induced by Inverse Stereographic Projection Shamal Chandra Karmaker In earlier studies of circular data, mostly circular distributions were considered and many biological data sets were assumed to be symmetric. However, presently inter- est has increased for skewed circular distributions as the assumption of symmetry may not be meaningful for some data. This thesis introduces three skewed circular models based on inverse stereographic projection, introduced by Minh and Farnum (2003), by considering three different versions of skewed-t considered in the litera- ture, namely Azzalini skewed-t, two-piece skewed-t and Jones and Faddy skewed-t. Shape properties of the resulting distributions along with estimation of parameters using maximum likelihood are discussed in this thesis.