An Automated Method to Enrich Consumer Health Vocabularies Using Glove Word Embeddings and an Auxiliary Lexical Resource
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Edical Ciences
INDIAN JOURNAL OF MEDICAL SCIENCES Indian Journal of Medical Sciences INDIAN JOURNAL VOLUME 62 ISSN 0019-5359 OF Indian Journal of Medical Sciences is a monthly journal published as a NUMBER 2 medium for the advancement of scientiÞ c knowledge in all the branches MEDICAL SCIENCES of Medicine and allied Sciences and publication of scientiÞ c investigation FEBRUARY 2008 in these Þ elds. It is also indented to present this as a form suitable to the general practitioner and primary care physician. ORIGINAL CONTRIBUTIONS Life time risk for development of ten major cancers in India and its trends over the years 1982 to 2000 The journal is owned by the Indian Journal of Medical Sciences Trust, a L. Satyanarayana, Smita asthana ........... ........... 35 Late Dr. J. C. Patel registered charitable organisation and published by Medknow Publications, Epidemiological correlates of nutritional anemia among children (6-35 months) in rural Wardha, Founder Editor Mumbai, India. central India Editor (1947-2003) N. Sinha, P. R. Deshmukh, B. S. Garg ........... ........... 45 The journal is indexed/listed with Index Medicus (Indian J Med Sci), Development of a scale for attitude toward condom use for migrant workers in India Arunansu Talukdar, Runa Bal, Debasis Sanyal, Krishnendu Roy, MEDLINE, PubMed, EMBASE, CAB Abstracts, Global Health, Health and Payel Sengupta Talukdar ........... ........... 55 Wellness Research Center, Health Reference Center Academic, InfoTrac One File, Expanded Academic ASAP, Journal Articles Database (JADE), CASE REPORTS Editor-in-Chief Dr. B. C. Mehta Indian Science Abstracts and PubList. Myasthenic crisis–like syndrome due to cleistanthus collinus poisoning Potikuri Damodaram, I. Chiranjeevi Manohar, D. Prabath Kumar, Alladi Mohan, Editor B. -

The Blue Book Standards for the Management and Evaluation of STI
GONORHEA CHLAMYDIA NGU MPC HIV HPV SYPHILIS YEAST SCABIES BACTERIAL VAGINOSIS TRICHOMONIASIS PUBIC LICE HERPES GONORHEA CHLAMYDIA NGU MPC HIV HPV SYPHILIS YEAST SCABIES BACTERIAL VAGINOSIS TRICHOMONIASIS PUBIC LICE HERPES GONORHEA CHLAMYDIA NGU MPC HIV HPV SYPHILIS YEAST SCABIES BACTERIAL VAGINOSIS TRICHOMONIASIS PUBIC LICE HERPES GONORHEA CHLAMYDIA NGU MPC HIV HPV SYPHILIS YEAST SCABIES BACTERIAL VAGINOSIS TRICHOMONIASIS PUBIC LICE HERPES The Blue Book Standards GONORHEA CHLAMYDIA NGU MPC HIV HPV SYPHILIS YEAST SCABIES for the Management and BACTERIAL VAGINOSIS TRICHOMONIASIS Evaluation of STI Clinic PUBIC LICE HERPES GONORHEA CHLAMYDIA NGU MPC HIV HPV Clients SYPHILIS YEAST SCABIES BACTERIAL VAGINOSIS TRICHOMONIASIS PUBIC LICE HERPES GONORHEA CHLAMYDIA NGU MPC HIV HPV SYPHILIS YEAST SCABIES BACTERIAL VAGINOSIS TRICHOMONIASIS PUBIC LICE HERPES GONORHEA CHLAMYDIA NGU MPC HIV HPV SYPHILIS YEAST SCABIES BACTERIAL VAGINOSIS TRICHOMONIASIS PUBIC LICE HERPES GONORHEA CHLAMYDIA NGU MPC HIV HPV SYPHILIS YEAST SCABIES BACTERIAL VAGINOSIS TRICHOMONIASIS Last Revised: June 2021 PUBIC LICE1 HERPES GONORHEA CHLAMYDIA NGU MPC HIV HPV YEAST pSYPHILIS YEAST SCABIES Acknowledgements A committee of provincial sexually transmitted disease representatives convened by Alberta Health Services (AHS) prepared this manual. Thanks are extended to the members of the STI Blue Book Working Group and the staff of the Alberta Provincial Laboratory for Public Health for their expertise and time to provide input and feedback in the development of the document. Special -
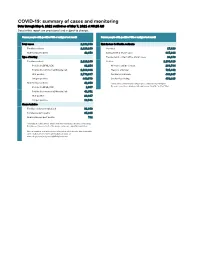
COVID-19: Summary of Cases and Monitoring Data Through May 6, 2021 Verified As of May 7, 2021 at 09:25 AM Data in This Report Are Provisional and Subject to Change
COVID-19: summary of cases and monitoring Data through May 6, 2021 verified as of May 7, 2021 at 09:25 AM Data in this report are provisional and subject to change. Cases: people with positive PCR or antigen test result Cases: people with positive PCR or antigen test result Total cases 2,262,598 Risk factors for Florida residents 2,220,240 Florida residents 2,220,240 Traveled 17,829 Non-Florida residents 42,358 Contact with a known case 887,914 Type of testing Traveled and contact with a known case 24,179 Florida residents 2,220,240 Neither 1,290,318 Positive by BPHL/CDC 81,254 No travel and no contact 268,784 Positive by commercial/hospital lab 2,138,986 Travel is unknown 725,442 PCR positive 1,770,667 Contact is unknown 493,047 Antigen positive 449,573 Contact is pending 459,315 Non-Florida residents 42,358 Travel can be unknown and contact can be unknown or pending for Positive by BPHL/CDC 1,007 the same case, these numbers will sum to more than the "neither" total. Positive by commercial/hospital lab 41,351 PCR positive 28,817 Antigen positive 13,541 Characteristics Florida residents hospitalized 91,848 Florida resident deaths 35,635 Non-Florida resident deaths 711 Hospitalized counts include anyone who was hospitalized at some point during their illness. It does not reflect the number of people currently hospitalized. More information on deaths identified through death certificate data is available on the National Center for Health Statistics website at www.cdc.gov/nchs/nvss/vsrr/COVID19/index.htm. -

Human Sexuality Ceu.Docx
1 Most people print off a copy of the post test and circle the answers as they read through the materials. Then, you can log in, go to "My Account" and under "Courses I Need to Take" click on the blue "Enter Answers" button. After completing the post test, you can print your certificate. Chapter 1 Topics in Human Sexuality: Sexuality Across the Lifespan Childhood and Adolescence Introduction Take a moment to think about your first sexual experience. Maybe it was an experience of early masturbation, “playing doctor” or “show me yours and I’ll show you mine.” Many of us do not think of childhood as a time of emerging sexuality, although we likely think of adolescence in just that way. But the assumption that sexuality begins in adolescence is incorrect. Human sexual development is a process that occurs throughout the lifespan. There are important biological and psychological aspects of sexuality that differ in children and adolescents, and later in adults and the elderly. In an early but seminal work on child sexuality Moglia & Knowles (1997) make a powerful statement. They state: from the moment they are born, infants are learning about their bodies, learning how to love and who to trust. Parents are their primary educators about sexuality, and mental health professionals often are called upon to guide these conversations. Like others who write about sexuality in childhood, these authors recognize that sexuality begins early and that parents play a pivotal role in educating children about their sexuality and about sexual expression. As clinicians working with families, it is important to be able to respond to questions regarding what is “normative” and to establish the foundation of healthy sexuality. -
Mischer Neuroscience Institute Report 2013
MISCHER NEUROSCIENCE INSTITUTE REPORT 2013 Built on a foundation of long-term collaboration between Memorial Hermann-Texas Medical Center, part of the 12-hospital Memorial Hermann Health System, and UTHealth Medical School, the Mischer Neuroscience Institute (MNI) is the largest and most comprehensive neuroscience program in Texas. The Institute brings together a team of world- class clinicians, researchers and educators whose insights and research findings are transforming the field of neuroscience. Thanks to their knowledge and talent, MNI is nationally recognized for leading-edge medicine and consistently ranked by quality benchmarking organizations as a leader in clinical quality and patient safety. Table of Contents 03 About the Institute 27 2013 Accolades 35 Scope of Services — Brain Tumor — Cerebrovascular — Children’s Neuroscience — Epilepsy — Movement Disorders — Multiple Sclerosis — Neurocognitive Disorders — Neuromuscular Disorders — Neurorehabilitation — Neurotrauma/Critical Care — Spine 76 Research & Innovation 102 Selected Publications 114 Patient Stories 131 Staff Listing Dear Esteemed Colleagues, 2013, our sixth anniversary, was a year of recognitions and strategic expansion at the Mischer Neuroscience Institute (MNI) at Memorial Hermann-Texas Medical Center and Mischer Neuroscience Associates (MNA), a citywide network of neurosurgeons and neurologists. Among those recognitions is the highly coveted Comprehensive Stroke Center certification awarded by The Joint Commission (TJC) and the American Heart Association/American Stroke Association. MNI is home to the first and only stroke program in Texas to meet TJC’s rigorous standards. While the neurosurgery program continues to grow, a major focus in 2013 was the expansion of neurology services across the city. Since 2008, MNI has worked neurosurgeons, neurologists and neurointensivists to extend its neurosurgery program from the Texas were welcomed to MNI and UTHealth. -

MEDICINE a Magazine for Alumni and Friends of the School of Medicine Volume 41 • Number 1 • Winter 2015
MEDICINE A Magazine for Alumni and Friends of the School of Medicine Volume 41 • Number 1 • Winter 2015 SCREENING METHOD: MLPA (MULTIPLEX LIGATION-DEPENDENT PROBE AMPLIFICA- TION/MRC HOLLAND, SALSA KIT PO34- PO345 VERSION A2 NEW PCR MIX): HYBRIDIZATION AND LIGATION/AMPLIFICA- TION OF SPECIFIC PROBES OF EACH OF THE 79 EXONS OF THE DMD GENE DP427 Frontiers OF THE FUTURE How Personalized Medicine,33% Genomics, and Informatics Are Transforming Health Care 26% 18% THIS INDIVIDUAL IS HETEROZYGOUS FOR THE C.1585-1G>A CF MUTA- TION. THE REMAINING MUTATIONS ARE ABSENT AND TOGETHER, THESE MUTATIONS ACCOUNT FOR APPROXI- MATELY 93.5% OF THE CF MUTATIONS SEEN IN THE LOCAL POPULATION. ANALYSIS BASED ON ALLELE-SPECIFIC dean’s view A Magazine for Alumni and Friends of the School of Medicine Volume 41 • Number 1 • Winter 2015 Tomorrow’s Medicine SCHOOL LEADERSHIP Senior Vice President and Dean Selwyn M. Vickers, M.D., FACS Executive Vice Dean This issue’s cover story is entitled “Frontiers of Anupam Agarwal, M.D. the Future,” but in many ways the future of health ALUMNI ASSOCIATION BOARD OF DIRECTORS President care is already here. As gene sequencing costs have Pink Lowe Folmar Jr., M.D. ’72 President-Elect plummeted due to advances in genomics and Timothy P. Hecker, M.D. ’04 Secretary/Treasurer computing technology, researchers are looking for Gerhard A. W. Boehm, M.D. ’71 Immediate Past President ways to use patients’ genomic data, along with Norman F. McGowin III, M.D. ’80 District Representatives J. Donald Kirby, M.D. ’72—1st District information about their lifestyle, behavior, and James H. -

2010 CAPHIS Top 100 List Health Websites You Can Trust GENERAL HEALTH Aetna Intelihealth
2010 CAPHIS Top 100 List Health Websites You Can Trust GENERAL HEALTH Aetna Intelihealth http://www.intelihealth.com/ Aetna InteliHealth, a subsidiary of Aetna, partners with Harvard Medical School and Columbia University College of Dental Medicine to provide health information on this website. It’s important to note that Aetna InteliHealth’s editorial policy states that it maintains absolute editorial independence from Aetna. That said, the site is content rich on disease and treatment information and includes nice additional features such as its Interactive Health Tools, Ask the Expert, Personalized Health E-mail service, and Discussion Boards. The Cleveland Clinic Health Information Center http://my.clevelandclinic.org/health/default.aspx Produced by the Cleveland Clinic Department of Patient Education and Health Information, this site offers information on over 900 health topics. Podcasts and webcasts of health information are available along with transcripts of web chats with physicians answering health questions. The Cleveland Clinic Patient Education and Health Information provides a live chat service Monday through Friday, 10:00 am to 1:30 pm EST (except holidays) where health educators are available to provide general health information and recommendations of web sites. Familydoctor.org http://familydoctor.org/online/famdocen/home.html This web site is offers clear and concise patient hand-outs for common medical concerns and conditions. All information has been written and reviewed by physicians and patient education professionals at the American Academy of Family Physicians. “Conditions A-Z”, make finding a hand-out easy. Handy buttons translates the site into Spanish and increases the type size. Other helpful features are a medical dictionary, health calculators, the Healthy Living Guides, a Smart Patient Guide, Health Tools section and a Health Tip of the Day. -
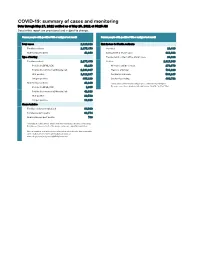
COVID-19: Summary of Cases and Monitoring Data Through May 27, 2021 Verified As of May 28, 2021 at 09:25 AM Data in This Report Are Provisional and Subject to Change
COVID-19: summary of cases and monitoring Data through May 27, 2021 verified as of May 28, 2021 at 09:25 AM Data in this report are provisional and subject to change. Cases: people with positive PCR or antigen test result Cases: people with positive PCR or antigen test result Total cases 2,320,818 Risk factors for Florida residents 2,277,470 Florida residents 2,277,470 Traveled 18,490 Non-Florida residents 43,348 Contact with a known case 916,492 Type of testing Traveled and contact with a known case 24,822 Florida residents 2,277,470 Neither 1,317,666 Positive by BPHL/CDC 83,133 No travel and no contact 278,679 Positive by commercial/hospital lab 2,194,337 Travel is unknown 733,229 PCR positive 1,812,287 Contact is unknown 508,245 Antigen positive 465,183 Contact is pending 460,702 Non-Florida residents 43,348 Travel can be unknown and contact can be unknown or pending for Positive by BPHL/CDC 1,035 the same case, these numbers will sum to more than the "neither" total. Positive by commercial/hospital lab 42,313 PCR positive 29,532 Antigen positive 13,816 Characteristics Florida residents hospitalized 94,930 Florida resident deaths 36,774 Non-Florida resident deaths 738 Hospitalized counts include anyone who was hospitalized at some point during their illness. It does not reflect the number of people currently hospitalized. More information on deaths identified through death certificate data is available on the National Center for Health Statistics website at www.cdc.gov/nchs/nvss/vsrr/COVID19/index.htm. -

An Automated Method to Enrich Consumer Health
Original Paper An Automated Method To Enrich Consumer Health Vocabularies Using GloVe Word Embeddings and An Auxiliary Lexical Resource Mohammed Ibrahim1*, PhD; Susan Gauch1*, PhD; Omar Salman1*, PhD; Mohammed Alqahatani1*, PhD 1Computer Science and Computer Engineering, University of Arkansas, Fayetteville, Arkansas, US *all authors contributed equally Abstract Background: Clear language makes communication easier between any two parties. A layman may have difficulty communicating with a professional due to not understanding the specialized terms common to the domain. In healthcare, it is rare to find a layman knowledgeable in medical jargon which can lead to poor understanding of their condition and/or treatment. To bridge this gap, several professional vocabularies and ontologies have been created to map laymen medical terms to professional medical terms and vice versa. Objective: Many of the presented vocabularies are built manually or semi- automatically requiring large investments of time and human effort and consequently the slow growth of these vocabularies. In this paper, we present an automatic method to enrich laymen's vocabularies that has the benefit of being able to be applied to vocabularies in any domain. Methods: Our entirely automatic approach uses machine learning, specifically Global Vectors for Word Embeddings (GloVe), on a corpus collected from a social media healthcare platform to extend and enhance consumer health vocabularies (CHV). Our approach further improves the CHV by incorporating synonyms and hyponyms from the WordNet ontology. The basic GloVe and our novel algorithms incorporating WordNet were evaluated using two laymen datasets from the National Library of Medicine (NLM), Open-Access Consumer Health Vocabulary (OAC CHV) and MedlinePlus Healthcare Vocabulary. -
Medical West Provider Directory
VM-5200002 VIVA MEDICARE Plus (HMO) VIVA MEDICARE Prime (HMO) VIVA MEDICARE Premier (HMO) VIVA MEDICARE Extra Value (HMO SNP) VIVA MEDICARE Select (HMO) PLAN PROVIDER DIRECTORY For the Medical West Provider System VIVA MEDICARE is an HMO plan with a Medicare contract and a contract with the Alabama Medicaid Agency. Enrollment in VIVA MEDICARE depends on contract renewal. This directory current as of Thursday, September 12, 2019 H0154_mcdoc2383A_C_08/23/2019 Page 1 of 192 VIVA MEDICARE Plus (HMO) VIVA MEDICARE Prime (HMO) VIVA MEDICARE Premier (HMO) VIVA MEDICARE Extra Value (HMO SNP) VIVA MEDICARE Select (HMO) PLAN PROVIDER DIRECTORY This directory provides a list of VIVA MEDICARE’s current network providers in the Medical West Provider System. This directory is for Autauga, Baldwin, Blount, Bullock, Calhoun, Cherokee, Chilton, Colbert, Crenshaw, Cullman, DeKalb, Dale, Elmore, Etowah, Franklin, Geneva, Henry, Houston Jefferson, Lauderdale, Lee, Lowndes, Macon, Mobile, Montgomery, Pike, Shelby, St. Clair, Talladega, Tallapoosa, and Walker Counties in Alabama. Not all plans offered by VIVA MEDICARE are available in each county listed above. See page 5 for the service area for each plan. To access VIVA MEDICARE’s online provider directory, you can visit www.VivaHealth.com/Medicare. For any questions about the information contained in this directory, please call our Member Services Department at 1-800-633-1542 toll free, Monday-Friday, 8 a.m. – 8 p.m. (October 1 – March 31, 7 days a week, 8 a.m. – 8 p.m.). TTY users should call 711. Member Services has free language interpreter services available for non-English speakers. This document may be available in alternate formats. -

Information Needs and Visitors' Experience of an Internet Expert
JOURNAL OF MEDICAL INTERNET RESEARCH Himmel et al Original Paper Information Needs and Visitors© Experience of an Internet Expert Forum on Infertility Wolfgang Himmel1, PhD; Juliane Meyer1, MD; Michael M Kochen1, MD, MPH, PhD, FRCGP; Hans Wilhelm Michelmann2, PhD 1Department of General Practice, University of Goettingen, Goettingen, Germany 2Department of Obstetrics and Gynaecology (Study Group of Reproductive Medicine), University of Goettingen, Goettingen, Germany Corresponding Author: Wolfgang Himmel, PhD Department of General Practice University of Goettingen Humboldtallee 38 37073 Goettingen Germany Phone: +49 0 551 392648 Fax: +49 0 551 399530 Email: [email protected] Abstract Background: Patients increasingly use health portals and Web-based expert forums (ask-the-doctor services), but little is known about the specific needs of Internet users visiting such websites, the nature of their requests, or how satisfied they are with Internet health experts. Objective: The aim of this study was to analyze the information requests of (mostly female) patients visiting an Internet expert forum on involuntary childlessness and their satisfaction with the experts© feedback. Methods: We posted an electronic questionnaire on a website hosting an expert forum on involuntary childlessness. The questionnaire was ªactivatedº whenever a visitor sent a question or request to the expert forum. The survey focused on the reasons for visiting the expert forum and whether the visitors were satisfied with the experts© answers to previously posted questions. The free-text questions of visitors who answered the survey were analyzed using Atlas-ti, a software program for qualitative data analysis. Results: Over a period of 6 months, 513 out of 610 visitors (84%) answered the questionnaire. -
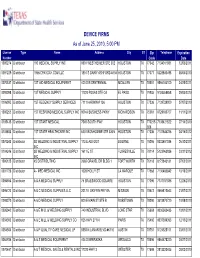
DEVICE FIRMS As of June 25, 2010, 5:00 PM
DEVICE FIRMS As of June 25, 2010, 5:00 PM License Type Name Address City ST Zip Telephone Expiration Number Code Date 1000274 Distributor 180 MEDICAL SUPPLY INC 9801 WESTHEIMER STE 302 HOUSTON TX 77042 7134011881 12/03/2011 0091229 Distributor 1866ICPAYDAY.