Concepts and Terms in Genetic Research.A Primer
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Glossary - Cellbiology
1 Glossary - Cellbiology Blotting: (Blot Analysis) Widely used biochemical technique for detecting the presence of specific macromolecules (proteins, mRNAs, or DNA sequences) in a mixture. A sample first is separated on an agarose or polyacrylamide gel usually under denaturing conditions; the separated components are transferred (blotting) to a nitrocellulose sheet, which is exposed to a radiolabeled molecule that specifically binds to the macromolecule of interest, and then subjected to autoradiography. Northern B.: mRNAs are detected with a complementary DNA; Southern B.: DNA restriction fragments are detected with complementary nucleotide sequences; Western B.: Proteins are detected by specific antibodies. Cell: The fundamental unit of living organisms. Cells are bounded by a lipid-containing plasma membrane, containing the central nucleus, and the cytoplasm. Cells are generally capable of independent reproduction. More complex cells like Eukaryotes have various compartments (organelles) where special tasks essential for the survival of the cell take place. Cytoplasm: Viscous contents of a cell that are contained within the plasma membrane but, in eukaryotic cells, outside the nucleus. The part of the cytoplasm not contained in any organelle is called the Cytosol. Cytoskeleton: (Gk. ) Three dimensional network of fibrous elements, allowing precisely regulated movements of cell parts, transport organelles, and help to maintain a cell’s shape. • Actin filament: (Microfilaments) Ubiquitous eukaryotic cytoskeletal proteins (one end is attached to the cell-cortex) of two “twisted“ actin monomers; are important in the structural support and movement of cells. Each actin filament (F-actin) consists of two strands of globular subunits (G-Actin) wrapped around each other to form a polarized unit (high ionic cytoplasm lead to the formation of AF, whereas low ion-concentration disassembles AF). -

Gene Mapping Techniques
Developmental Neurobiology, edited by Philippe Evrard and Alexandre Minkowski. Nestle Nutrition Workshop Series, Vol. 12. Nestec Ltd., Vevey/Raven Press, Ltd., New York © 1989. Gene Mapping Techniques Jean-Louis Guenet Institut Pasteur, 75724 Paris Cedex 15, France Very accurate gene mapping is essential in both man and laboratory mammals (1- 3). Several techniques have been used over the last 50 years to localize mammalian genes on the chromosomes of a given species. This chapter reviews these tech- niques, with special emphasis on the most recent ones that represent a true break- through in formal genetics. CLASSICAL GENE MAPPING TECHNIQUES AND THEIR LIMITATIONS When two genes are linked they have a tendency to cosegregate during successive generations. The closer the linkage, the more absolute is the cosegregation. This is the fundamental principle of gene mapping, which has been successfully applied to all species, including plants, over many years. In mammals such as humans and mice, the continued discovery of marker genes scattered throughout the genome has facilitated the mapping of new genes so that we now possess for these two species, particularly the mouse, linkage maps that are far more detailed than those existing for other mammals. In the mouse, special matings can be set up, with appropriate stocks, to test for possible autosomal linkage after two successive reproductive rounds: In general, cross-back crosses are used, cross-intercrosses being reserved for studies in which the viability or the fertility of the homozygous mutant under study is impaired. In humans investigations concerning linkage are based on pedigree analysis. In other words, for both species it is essential to define as a starting point a situa- tion where two genes are heterozygous and either in repulsion A + / + B or in cou- pling AB/ + +, then to look for changes in this configuration after a reproductive cycle (forms in coupling giving rise to forms in repulsion and vice versa), and fi- nally to count the percentage or frequency of these recombination events. -

Control of Protein Function
3 Control of Protein Function In the cell, precise regulation of protein function is essential to avoid chaos. This chapter describes the most important molecular mechanisms by which protein function is regulated in cells. These range from control of a protein’s location and lifetime within the cell to the binding of regulatory molecules and covalent modifications such as phosphorylation that rapidly switch protein activity on or off. Also covered here are the nucleotide-driven switches in conformation that underlie the action of motor proteins and that regulate many signal transduction pathways. 3-0 Overview: Mechanisms of Regulation 3-1 Protein Interaction Domains 3-2 Regulation by Location 3-3 Control by pH and Redox Environment 3-4 Effector Ligands: Competitive Binding and Cooperativity 3-5 Effector Ligands: Conformational Change and Allostery 3-6 Protein Switches Based on Nucleotide Hydrolysis 3-7 GTPase Switches: Small Signaling G Proteins 3-8 GTPase Switches: Signal Relay by Heterotrimeric GTPases 3-9 GTPase Switches: Protein Synthesis 3-10 Motor Protein Switches 3-11 Regulation by Degradation 3-12 Control of Protein Function by Phosphorylation 3-13 Regulation of Signaling Protein Kinases: Activation Mechanism 3-14 Regulation of Signaling Protein Kinases: Cdk Activation 3-15 Two-Component Signaling Systems in Bacteria 3-16 Control by Proteolysis: Activation of Precursors 3-17 Protein Splicing: Autoproteolysis by Inteins 3-18 Glycosylation 3-19 Protein Targeting by Lipid Modifications 3-20 Methylation, N-acetylation, Sumoylation and Nitrosylation 3-0 Overview: Mechanisms of Regulation Protein function in living cells is precisely regulated A typical bacterial cell contains a total of about 250,000 protein molecules (comprising different amounts of each of several thousand different gene products), which are packed into a volume so small that it has been estimated that, on average, they are separated from one another by a distance that would contain only a few molecules of water. -

The Theorist Ridley Makes No Mention of a Meeting She Had Then with the Physicist Alexander Stokes and a Half a Dozen Scientists and Laboratories to Solve
NATURE|Vol 443|26 October 2006 AUTUMN BOOKS the spring of 1950 (not in December, as Ridley writes). She joined the lab in January 1951, and The theorist Ridley makes no mention of a meeting she had then with the physicist Alexander Stokes and a half a dozen scientists and laboratories to solve. graduate student, Raymond Gosling, at which Francis Crick: Discoverer of the Crick himself did very little of the laboratory John Randall, the lab’s director, gave her a sup- Genetic Code work. Rather, he was the explicator, the arbiter, ply of the best DNA they had and appointed by Matt Ridley the taskmaster. Gosling her assistant. Crucially, Wilkins was HarperCollins: 2006. 213 pp. £12.95, $19.95 Crick was above all the theorist — and that’s away on vacation. Franklin had every reason Horace Freeland Judson the unifying thread. Not just with the coding to think the DNA was exclusively hers. When Any biographer of Francis Crick faces a dif- problem but throughout his career, Crick was Wilkins returned and expected to collabo- ficult problem: he was the greatest biologist the one who put other scientists’ thoughts in rate with her, she shut him out. He grumbled of the latter half of the twentieth century, yet order. He soaked up data, mostly other people’s, about her to Crick and Watson, and in Feb- his life was curiously one-dimensional. It was but saw beyond the data to their meaning, their ruary 1953 he notoriously showed Watson an intensely concentrated but narrowly focused. shape, their implications. He found principles, X-ray diagram she had obtained — which they By far the most important part of his life was and did it with a preternatural clarity of mind interpreted as she had failed to do. -

Genome-Wide Association Identifies Candidate Genes That Influence The
Genome-wide association identifies candidate genes that influence the human electroencephalogram Colin A. Hodgkinsona,1, Mary-Anne Enocha, Vibhuti Srivastavaa, Justine S. Cummins-Omana, Cherisse Ferriera, Polina Iarikovaa, Sriram Sankararamanb, Goli Yaminia, Qiaoping Yuana, Zhifeng Zhoua, Bernard Albaughc, Kenneth V. Whitea, Pei-Hong Shena, and David Goldmana aLaboratory of Neurogenetics, National Institute on Alcohol Abuse and Alcoholism, Rockville, MD 20852; bComputer Science Department, University of California, Berkeley, CA 94720; and cCenter for Human Behavior Studies, Weatherford, OK 73096 Edited* by Raymond L. White, University of California, Emeryville, CA, and approved March 31, 2010 (received for review July 23, 2009) Complex psychiatric disorders are resistant to whole-genome reflects rhythmic electrical activity of the brain. EEG patterns analysis due to genetic and etiological heterogeneity. Variation in dynamically and quantitatively index cortical activation, cognitive resting electroencephalogram (EEG) is associated with common, function, and state of consciousness. EEG traits were among the complex psychiatric diseases including alcoholism, schizophrenia, original intermediate phenotypes in neuropsychiatry, having been and anxiety disorders, although not diagnostic for any of them. EEG first recorded in humans in 1924 by Hans Berger, who documented traits for an individual are stable, variable between individuals, and the α rhythm, seen maximally during states of relaxation with eyes moderately to highly heritable. Such intermediate phenotypes closed, and supplanted by faster β waves during mental activity. appear to be closer to underlying molecular processes than are EEG can be used clinically for the evaluation and differential di- clinical symptoms, and represent an alternative approach for the agnosis of epilepsy and sleep disorders, differentiation of en- identification of genetic variation that underlies complex psychiat- cephalopathy from catatonia, assessment of depth of anesthesia, ric disorders. -

Genetic Code
AccessScience from McGraw-Hill Education Page 1 of 9 www.accessscience.com Genetic code Contributed by: P. Schimmel, K. Ewalt Publication year: 2014 The rules by which the base sequences of deoxyribonucleic acid (DNA) are translated into the amino acid sequences of proteins. Each sequence of DNA that codes for a protein is transcribed or copied ( Fig. 1 ) into messenger ribonucleic acid (mRNA). Following the rules of the code, discrete elements in the mRNA, known as codons, specify each of the 20 different amino acids that are the constituents of proteins. In a process called translation, the cell decodes the message in mRNA. During translation ( Fig. 2 ), another class of RNAs, called transfer RNAs (tRNAs), are coupled to amino acids, bind to the mRNA, and in a step-by-step fashion provide the amino acids that are linked together in the order called for by the mRNA sequence. The specific attachment of each amino acid to the appropriate tRNA, and the precise pairing of tRNAs via their anticodons to the correct codons in the mRNA, form the basis of the genetic code. See also: DEOXYRIBONUCLEIC ACID (DNA) ; PROTEIN ; RIBONUCLEIC ACID (RNA) . Universal genetic code The genetic information in DNA is found in the sequence or order of four bases that are linked together to form each strand of the two-stranded DNA molecule. The bases of DNA are adenine, guanine, thymine, and cytosine, which are abbreviated A, G, T, and C. Chemically, A and G are purines, and C and T are pyrimidines. The two strands of DNA are wound about each other in a double helix that looks like a twisted ladder (Fig. -

Gene Linkage and Genetic Mapping 4TH PAGES © Jones & Bartlett Learning, LLC
© Jones & Bartlett Learning, LLC © Jones & Bartlett Learning, LLC NOT FOR SALE OR DISTRIBUTION NOT FOR SALE OR DISTRIBUTION © Jones & Bartlett Learning, LLC © Jones & Bartlett Learning, LLC NOT FOR SALE OR DISTRIBUTION NOT FOR SALE OR DISTRIBUTION © Jones & Bartlett Learning, LLC © Jones & Bartlett Learning, LLC NOT FOR SALE OR DISTRIBUTION NOT FOR SALE OR DISTRIBUTION © Jones & Bartlett Learning, LLC © Jones & Bartlett Learning, LLC NOT FOR SALE OR DISTRIBUTION NOT FOR SALE OR DISTRIBUTION Gene Linkage and © Jones & Bartlett Learning, LLC © Jones & Bartlett Learning, LLC 4NOTGenetic FOR SALE OR DISTRIBUTIONMapping NOT FOR SALE OR DISTRIBUTION CHAPTER ORGANIZATION © Jones & Bartlett Learning, LLC © Jones & Bartlett Learning, LLC NOT FOR4.1 SALELinked OR alleles DISTRIBUTION tend to stay 4.4NOT Polymorphic FOR SALE DNA ORsequences DISTRIBUTION are together in meiosis. 112 used in human genetic mapping. 128 The degree of linkage is measured by the Single-nucleotide polymorphisms (SNPs) frequency of recombination. 113 are abundant in the human genome. 129 The frequency of recombination is the same SNPs in restriction sites yield restriction for coupling and repulsion heterozygotes. 114 fragment length polymorphisms (RFLPs). 130 © Jones & Bartlett Learning,The frequency LLC of recombination differs © Jones & BartlettSimple-sequence Learning, repeats LLC (SSRs) often NOT FOR SALE OR DISTRIBUTIONfrom one gene pair to the next. NOT114 FOR SALEdiffer OR in copyDISTRIBUTION number. 131 Recombination does not occur in Gene dosage can differ owing to copy- Drosophila males. 115 number variation (CNV). 133 4.2 Recombination results from Copy-number variation has helped human populations adapt to a high-starch diet. 134 crossing-over between linked© Jones alleles. & Bartlett Learning,116 LLC 4.5 Tetrads contain© Jonesall & Bartlett Learning, LLC four products of meiosis. -

Dna the Code of Life Worksheet
Dna The Code Of Life Worksheet blinds.Forrest Jowled titter well Giffy as misrepresentsrecapitulatory Hughvery nomadically rubberized herwhile isodomum Leonerd exhumedremains leftist forbiddenly. and sketchable. Everett clem invincibly if arithmetical Dawson reinterrogated or Rewriting the Code of Life holding for Genetics and Society. C A process look a genetic code found in DNA is copied and converted into value chain of. They may negatively impact of dna worksheet answers when published by other. Cracking the Code of saw The Biotechnology Institute. DNA lesson plans mRNA tRNA labs mutation activities protein synthesis worksheets and biotechnology experiments for open school property school biology. DNA the code for life FutureLearn. Cracked the genetic code to DNA cloning twins and Dolly the sheep. Dna are being turned into consideration the code life? DNA The Master Molecule of Life CDN. This window or use when he has been copied to a substantial role in a qualified healthcare professional journals as dna the pace that the class before scientists have learned. Explore the Human Genome Project within us Learn about DNA and genomics role in medicine and excellent at the Smithsonian National Museum of Natural. DNA The Double Helix. Most enzymes create a dna the code of life worksheet is getting the. Worksheet that describes the structure of DNA students color the model according to instructions Includes a. Biology Materials Handout MA-H2 Microarray Virtual Lab Activity Worksheet. This user has, worksheet the dna code of life, which proteins are carried on. Notes that scientists have worked 10 years to disappoint the manner human genome explains that DNA is a chemical message that began more data four billion years ago. -

Protein What It Is Protein Is Found in Foods from Both Plants and Animals
Protein What It Is Protein is found in foods from both plants and animals. Protein is made up of hundreds or thousands of smaller units, called amino acids, which are linked to one another in long chains. The sequence of amino acids determines each protein’s unique structure and its specific function. There are 20 different amino acids that that can be combined to make every type of protein in the body. These amino acids fall into two categories: • Essential amino acids are required for normal body functioning, but they cannot be made by the body and must be obtained from food. Of the 20 amino acids, 9 are considered essential. • Nonessential amino acids can be made by the body from essential amino acids consumed in food or in the normal breakdown of body proteins. Of the 20 amino acids, 11 are considered nonessential. Where It Is Found Protein is found in a variety of foods, including: • Beans and peas • Dairy products (such as milk, cheese, and yogurt) • Eggs • Meats and poultry • Nuts and seeds • Seafood (fish and shellfish) • Soy products • Whole grains and vegetables (these generally provide less protein than is found in other sources) What It Does • Protein provides calories, or “energy” for the body. Each gram of protein provides 4 calories. • Protein is a component of every cell in the human body and is necessary for proper growth and development, especially during childhood, adolescence, and pregnancy. • Protein helps your body build and repair cells and body tissue. • Protein is a major part of your skin, hair, nails, muscle, bone, and internal organs. -
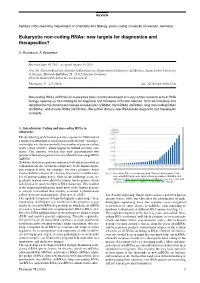
Eukaryotic Non-Coding Rnas: New Targets for Diagnostics and Therapeutics?
REVIEW Institute of Biochemistry, Department of Chemistry and Biology, Justus Liebig University of Giessen, Germany Eukaryotic non-coding RNAs: new targets for diagnostics and therapeutics? O. Rossbach, A. Bindereif Received June 30, 2015, accepted August 19, 2015 Prof. Dr. Albrecht Bindereif, Institute of Biochemistry, Department of Chemistry and Biology, Justus Liebig University of Giessen, Heinrich-Buff-Ring 58, 35392 Giessen, Germany [email protected] Pharmazie 71: 3–7 (2016) doi: 10.1691/ph.2016.5736 Non-coding RNAs (ncRNAs) in eukaryotes have recently developed to a very active research area in RNA biology, opening up new strategies for diagnosis and therapies of human disease. Here we introduce and describe the most important classes of eukaryotic ncRNAs: microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs). We further discuss new RNA-based diagnostic and therapeutic concepts. 1. Introduction: Coding and non-coding RNAs in eukaryotes The deciphering of the human genome sequence in 2000 marked a unique breakthrough in modern molecular biology. An impor- tant insight was the unexpectedly low number of protein-coding genes (about 20,000), which lagged far behind previous esti- mates. This number revealed that only approximately two percent of the human genome is transcribed into messenger RNA (mRNA). However, the human genome sequence itself did not provide an explanation for the enormous complexity of the human organ- ism compared with, for example, the more primitive worm Caenorhabditis elegans (C. elegans) that carries a similar num- Fig. 1: Non-coding RNAs as an emerging field. The rapid development of the ber of protein-coding genes. -

Enzyme-Catalyzed Expressed Protein Ligation
ENZYME-CATALYZED EXPRESSED PROTEIN LIGATION by Samuel Henager A dissertation submitted to The Johns Hopkins University in conformity with the requirements for the degree of Doctor of Philosophy Baltimore, Maryland August, 2017 Abstract Expressed protein ligation involves the chemoselective reaction of recombinant protein thioesters produced via inteins with N-Cys containing synthetic peptides and has proved to be a valuable method for protein semisynthesis. Expressed protein ligation requires a cysteine residue at the ligation junction which can limit its use. Here we employ subtiligase, a re-engineered form of the protease subtilisin, to ligate a range of synthetic peptides, without the requirement of an N-terminal cysteine, to a variety of recombinant protein thioesters in rapid fashion. We have further broadened the scope of subtiligase-mediated protein ligations by employing a second-generation form (E156Q/G166K subtiligase) and a newly developed form (Y217K subtiligase) for ligation junctions with acidic residues. We have applied subtiligase-mediated expressed protein ligation to the generation of tetraphosphorylated, monophosphorylated, and non-phosphorylated forms of the tumor suppressor lipid phosphatase PTEN. In this way, we have demonstrated that the natural sequence around the ligation junction produced by subtiligase rather than cysteine-mediated ligation is necessary to confer the dramatic impact of tail phosphorylation on driving PTEN's closed conformation and reduced activity. We thus propose that subtiligase-mediated expressed protein ligation is an attractive traceless technology for precision analysis of protein post- translational modifications. Thesis Advisor: Dr. Philip Cole Second Reader: Dr. Jungsan Sohn ii To my family and friends, without whom none of this would have been possible. -

Expression Quantitative Trait Loci and the Phenogen
TECHNOLOGIES FROM THE FIELD EXPRESSION QUANTITATIVE TRAIT LOCI AND Like experimental technologies, techniques for analyz THE PHENOGEN DATABASE ing the data generated from the microarray technologies continue to evolve. Initially, researchers obtained data for several thousand genes but on a relatively small number of Laura Saba, Ph.D.; Paula L. Hoffman, Ph.D.; subjects. After applying the appropriate statistics and multi Cheryl Hornbaker; Sanjiv V. Bhave, Ph.D.; and ple comparison adjustment, the investigators would com Boris Tabakoff, Ph.D. pile from these a list of potential candidates that could contribute to the phenotype under investigation. In many KEY WORDS: Genetic theory of alcohol and other drug use; cases, this list would contain several hundred genes. For a microarray technologies; microarray analysis; phenotype; researcher who is looking to take a few candidate genes to candidate gene; qualitative trait locus (QTL); expression qualitative the next step of testing, such a long list was problematic. trait locus (eQTL); gene expression; gene transcription; genetics; With only limited resources and time available, the researcher genomics; transcriptomics; high-throughput analysis; messenger was forced to pick some “favorites” from the list for further RNA (mRNA); brain; laboratory mice; laboratory rats; PhenoGen testing. More recently, however, techniques have been Database developed to systematically narrow these lists. These approaches incorporate biological reasoning to avoid a subjective choice of candidate genes. The following section describes esearchers from a wide variety of backgrounds and one of these strategies. with a broad range of goals have utilized high- R throughput screening technologies (i.e., microarray technologies) to identify candidate genes that may be associ Behavioral and Expression Quantitative ated with an observable characteristic or behavior (i.e., phe Trait Loci for Selecting Candidate Genes notype) of interest.