NIH Public Access Author Manuscript Dev Disabil Res Rev
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplementary Materials: Evaluation of Cytotoxicity and Α-Glucosidase Inhibitory Activity of Amide and Polyamino-Derivatives of Lupane Triterpenoids
Supplementary Materials: Evaluation of cytotoxicity and α-glucosidase inhibitory activity of amide and polyamino-derivatives of lupane triterpenoids Oxana B. Kazakova1*, Gul'nara V. Giniyatullina1, Akhat G. Mustafin1, Denis A. Babkov2, Elena V. Sokolova2, Alexander A. Spasov2* 1Ufa Institute of Chemistry of the Ufa Federal Research Centre of the Russian Academy of Sciences, 71, pr. Oktyabrya, 450054 Ufa, Russian Federation 2Scientific Center for Innovative Drugs, Volgograd State Medical University, Novorossiyskaya st. 39, Volgograd 400087, Russian Federation Correspondence Prof. Dr. Oxana B. Kazakova Ufa Institute of Chemistry of the Ufa Federal Research Centre of the Russian Academy of Sciences 71 Prospeсt Oktyabrya Ufa, 450054 Russian Federation E-mail: [email protected] Prof. Dr. Alexander A. Spasov Scientific Center for Innovative Drugs of the Volgograd State Medical University 39 Novorossiyskaya st. Volgograd, 400087 Russian Federation E-mail: [email protected] Figure S1. 1H and 13C of compound 2. H NH N H O H O H 2 2 Figure S2. 1H and 13C of compound 4. NH2 O H O H CH3 O O H H3C O H 4 3 Figure S3. Anticancer screening data of compound 2 at single dose assay 4 Figure S4. Anticancer screening data of compound 7 at single dose assay 5 Figure S5. Anticancer screening data of compound 8 at single dose assay 6 Figure S6. Anticancer screening data of compound 9 at single dose assay 7 Figure S7. Anticancer screening data of compound 12 at single dose assay 8 Figure S8. Anticancer screening data of compound 13 at single dose assay 9 Figure S9. Anticancer screening data of compound 14 at single dose assay 10 Figure S10. -

Yeast As a Model Organism to Study Diseases of Protein Misfolding
Tiago Fleming de Oliveira Outeiro YEAST AS A MODEL ORGANISM TO STUDY DISEASES OF PROTEIN MISFOLDING Instituto de Ciências Biomédicas de Abel Salazar Universidade do Porto Porto 2004 Tiago Fleming de Oliveira Outeiro YEAST AS A MODEL ORGANISM TO STUDY DISEASES OF PROTEIN MISFOLDING Dissertação de candidatura ao Grau de Doutor em Ciências Biomédicas submetida ao Instituto de Ciências Biomédicas de Abel Salazar. Orientadora - Professora Susan Lindquist Co-orientadora - Professora Maria João Saraiva Para os devidos efeitos, e de acordo com o disposto no n° 2 do Artigo 8 do Decreto-Lei n° 388/70, o autor desta dissertação declara que interveio na concepção e execução do trabalho experimental, na interpretação e discussão dos resultados e na preparação dos manuscriptos publicados e em vias de publicação. Na presente dissertação incluem-se os resultados das seguintes publicações: Outeiro, TF, Lindquist, S, (2003) Yeast cells provide insight into alpha-synuclein biology and pathobiology, Science, 302:1772-5. Willingham S, Outeiro TF, DeVit MJ, Lindquist SL, Muchowski PJ (2003) Yeast genes that enhance the toxicity of a mutant huntingtin fragment or alpha- synuclein, Science, 302:1769-72. Outeiro, TF, et al., Drug screening yields new insight into neurodegeneration, Nature, in press. Table of Contents TABLE OF CONTENTS ACKNOWLEDGEMENTS XIII ABSTRACT XVII RESUMO XIX OVERVIEW XXIII ORGANIZATION OF THE THESIS XXV ABBREVIATIONS XXVII CHAPTER 1. GENERAL INTRODUCTION 3 1.1 PROTEIN FOLDING AND MISFOLDING 3 1.2 CELLULAR QUALITY CONTROL MECHANISMS 6 1.2.1 -

A Large-Scale Overexpression Screen in Saccharomyces Cerevisiae Identifies Previously Uncharacterized Cell Cycle Genes
A large-scale overexpression screen in Saccharomyces cerevisiae identifies previously uncharacterized cell cycle genes Lauren F. Stevenson*, Brian K. Kennedy, and Ed Harlow Massachusetts General Hospital Cancer Center, Building 149, 13th Street, Charlestown, MA 02129 Contributed by Ed Harlow, January 5, 2001 We have undertaken an extensive screen to identify Saccharomyces in some, if not many, instances, effects on the cell cycle might be cerevisiae genes whose products are involved in cell cycle progres- apparent in the absence of complete lethality. For this reason, we sion. We report the identification of 113 genes, including 19 hypo- devised a protocol that would uncover not only those genes whose thetical ORFs, which confer arrest or delay in specific compartments overproduction is lethal, but also those where overproduction of the cell cycle when overexpressed. The collection of genes identi- causes impaired growth. We also reasoned that moderate overpro- fied by this screen overlaps with those identified in loss-of-function duction of proteins might be more physiologically relevant than cdc screens but also includes genes whose products have not previ- dramatic overproduction, therefore we used GAL promoter-driven ously been implicated in cell cycle control. Through analysis of strains libraries expressed from ARS-CEN vectors to control levels of gene lacking these hypothetical ORFs, we have identified a variety of new expression. CDC and checkpoint genes. Materials and Methods ell cycle studies performed with Saccharomyces cerevisiae have Screening of Libraries. Yeast strain K699 (W303 background) was Cserved as a guideline for understanding eukaryotic cell cycle transformed as previously described (14) with the cDNA library or progression. -

Metabolic Targets of Coenzyme Q10 in Mitochondria
antioxidants Review Metabolic Targets of Coenzyme Q10 in Mitochondria Agustín Hidalgo-Gutiérrez 1,2,*, Pilar González-García 1,2, María Elena Díaz-Casado 1,2, Eliana Barriocanal-Casado 1,2, Sergio López-Herrador 1,2, Catarina M. Quinzii 3 and Luis C. López 1,2,* 1 Departamento de Fisiología, Facultad de Medicina, Universidad de Granada, 18016 Granada, Spain; [email protected] (P.G.-G.); [email protected] (M.E.D.-C.); [email protected] (E.B.-C.); [email protected] (S.L.-H.) 2 Centro de Investigación Biomédica, Instituto de Biotecnología, Universidad de Granada, 18016 Granada, Spain 3 Department of Neurology, Columbia University Medical Center, New York, NY 10032, USA; [email protected] * Correspondence: [email protected] (A.H.-G.); [email protected] (L.C.L.); Tel.: +34-958-241-000 (ext. 20197) (L.C.L.) Abstract: Coenzyme Q10 (CoQ10) is classically viewed as an important endogenous antioxidant and key component of the mitochondrial respiratory chain. For this second function, CoQ molecules seem to be dynamically segmented in a pool attached and engulfed by the super-complexes I + III, and a free pool available for complex II or any other mitochondrial enzyme that uses CoQ as a cofactor. This CoQ-free pool is, therefore, used by enzymes that link the mitochondrial respiratory chain to other pathways, such as the pyrimidine de novo biosynthesis, fatty acid β-oxidation and amino acid catabolism, glycine metabolism, proline, glyoxylate and arginine metabolism, and sulfide oxidation Citation: Hidalgo-Gutiérrez, A.; metabolism. Some of these mitochondrial pathways are also connected to metabolic pathways González-García, P.; Díaz-Casado, in other compartments of the cell and, consequently, CoQ could indirectly modulate metabolic M.E.; Barriocanal-Casado, E.; López-Herrador, S.; Quinzii, C.M.; pathways located outside the mitochondria. -

Human Mitochondrial Pathologies of the Respiratory Chain and ATP Synthase: Contributions from Studies of Saccharomyces Cerevisiae
life Review Human Mitochondrial Pathologies of the Respiratory Chain and ATP Synthase: Contributions from Studies of Saccharomyces cerevisiae Leticia V. R. Franco 1,2,* , Luca Bremner 1 and Mario H. Barros 2 1 Department of Biological Sciences, Columbia University, New York, NY 10027, USA; [email protected] 2 Department of Microbiology,Institute of Biomedical Sciences, Universidade de Sao Paulo, Sao Paulo 05508-900, Brazil; [email protected] * Correspondence: [email protected] Received: 27 October 2020; Accepted: 19 November 2020; Published: 23 November 2020 Abstract: The ease with which the unicellular yeast Saccharomyces cerevisiae can be manipulated genetically and biochemically has established this organism as a good model for the study of human mitochondrial diseases. The combined use of biochemical and molecular genetic tools has been instrumental in elucidating the functions of numerous yeast nuclear gene products with human homologs that affect a large number of metabolic and biological processes, including those housed in mitochondria. These include structural and catalytic subunits of enzymes and protein factors that impinge on the biogenesis of the respiratory chain. This article will review what is currently known about the genetics and clinical phenotypes of mitochondrial diseases of the respiratory chain and ATP synthase, with special emphasis on the contribution of information gained from pet mutants with mutations in nuclear genes that impair mitochondrial respiration. Our intent is to provide the yeast mitochondrial specialist with basic knowledge of human mitochondrial pathologies and the human specialist with information on how genes that directly and indirectly affect respiration were identified and characterized in yeast. Keywords: mitochondrial diseases; respiratory chain; yeast; Saccharomyces cerevisiae; pet mutants 1. -

Protein Moonlighting Revealed by Non-Catalytic Phenotypes of Yeast Enzymes
Genetics: Early Online, published on November 10, 2017 as 10.1534/genetics.117.300377 Protein Moonlighting Revealed by Non-Catalytic Phenotypes of Yeast Enzymes Adriana Espinosa-Cantú1, Diana Ascencio1, Selene Herrera-Basurto1, Jiewei Xu2, Assen Roguev2, Nevan J. Krogan2 & Alexander DeLuna1,* 1 Unidad de Genómica Avanzada (Langebio), Centro de Investigación y de Estudios Avanzados del IPN, 36821 Irapuato, Guanajuato, Mexico. 2 Department of Cellular and Molecular Pharmacology, University of California, San Francisco, San Francisco, California, 94158, USA. *Corresponding author: [email protected] Running title: Genetic Screen for Moonlighting Enzymes Keywords: Protein moonlighting; Systems genetics; Pleiotropy; Phenotype; Metabolism; Amino acid biosynthesis; Saccharomyces cerevisiae 1 Copyright 2017. 1 ABSTRACT 2 A single gene can partake in several biological processes, and therefore gene 3 deletions can lead to different—sometimes unexpected—phenotypes. However, it 4 is not always clear whether such pleiotropy reflects the loss of a unique molecular 5 activity involved in different processes or the loss of a multifunctional protein. Here, 6 using Saccharomyces cerevisiae metabolism as a model, we systematically test 7 the null hypothesis that enzyme phenotypes depend on a single annotated 8 molecular function, namely their catalysis. We screened a set of carefully selected 9 genes by quantifying the contribution of catalysis to gene-deletion phenotypes 10 under different environmental conditions. While most phenotypes were explained 11 by loss of catalysis, slow growth was readily rescued by a catalytically-inactive 12 protein in about one third of the enzymes tested. Such non-catalytic phenotypes 13 were frequent in the Alt1 and Bat2 transaminases and in the isoleucine/valine- 14 biosynthetic enzymes Ilv1 and Ilv2, suggesting novel "moonlighting" activities in 15 these proteins. -

COQ2 Gene Coenzyme Q2, Polyprenyltransferase
COQ2 gene coenzyme Q2, polyprenyltransferase Normal Function The COQ2 gene provides instructions for making an enzyme that carries out one step in the production of a molecule called coenzyme Q10, which has several critical functions in cells throughout the body. In cell structures called mitochondria, coenzyme Q10 plays an essential role in a process called oxidative phosphorylation, which converts the energy from food into a form cells can use. Coenzyme Q10 is also involved in producing pyrimidines, which are building blocks of DNA, its chemical cousin RNA, and molecules such as ATP and GTP that serve as energy sources in the cell. In cell membranes, coenzyme Q10 acts as an antioxidant, protecting cells from damage caused by unstable oxygen-containing molecules (free radicals), which are byproducts of energy production. Health Conditions Related to Genetic Changes Multiple system atrophy Several variations in the COQ2 gene have been suggested to increase the risk of multiple system atrophy, a progressive brain disorder that affects movement and balance and disrupts the function of the autonomic nervous system. The autonomic nervous system controls body functions that are mostly involuntary, such as regulation of blood pressure. The identified variations alter single protein building blocks (amino acids) in the COQ2 enzyme. Most of the variations are very rare, but a genetic change that replaces the amino acid valine with the amino acid alanine at position 393 (written as Val393Ala or V393A) is relatively common. Studies suggest that these variations, including V393A, are associated with an increased risk of developing multiple system atrophy in the Japanese population. However, studies have not found a correlation between COQ2 gene variations and multiple system atrophy in other populations, including Koreans, Europeans, and North Americans. -

The Yeast Deletion Collection: a Decade of Functional Genomics
YEASTBOOK CELL CYCLE The Yeast Deletion Collection: A Decade of Functional Genomics Guri Giaever and Corey Nislow University of British Columbia, Vancouver, British Columbia, Canada V6T1Z3 ABSTRACT The yeast deletion collections comprise .21,000 mutant strains that carry precise start-to-stop deletions of 6000 open reading frames. This collection includes heterozygous and homozygous diploids, and haploids of both MATa and MATa mating types. The yeast deletion collection, or yeast knockout (YKO) set, represents the first and only complete, systematically constructed deletion collection available for any organism. Conceived during the Saccharomyces cerevisiae sequencing project, work on the project began in 1998 and was completed in 2002. The YKO strains have been used in numerous laboratories in .1000 genome-wide screens. This landmark genome project has inspired development of numerous genome-wide technologies in organisms from yeast to man. Notable spinoff technologies include synthetic genetic array and HIPHOP chemogenomics. In this retrospective, we briefly describe the yeast deletion project and some of its most noteworthy biological contributions and the impact that these collections have had on the yeast research community and on genomics in general. TABLE OF CONTENTS Abstract 451 Yeast as a Model for Molecular Genetics 452 A Brief History of the Saccharomyces Genome Deletion Project 452 Managing the Collection: Cautions and Caveats 453 Early Applications of the Deletion Collection 455 Seminal publications 455 Genome-wide phenotypic screens 456 Early genome-wide screens 458 Comparing genome-wide studies between laboratories: 458 Mitochondrial respiration as a case study: 458 Metrics to assess deletion strain fitness: 458 Large-Scale Phenotypic Screens 459 Cell growth 459 Mating, sporulation, and germination 459 Membrane trafficking 460 Selected environmental stresses 460 Continued Copyright © 2014 by the Genetics Society of America doi: 10.1534/genetics.114.161620 Available freely online through the author-supported open access option. -

Systems Cell Biology of the Mitotic Spindle
JCB: Comment Systems cell biology of the mitotic spindle Ramsey A. Saleem and John D. Aitchison Institute for Systems Biology, Seattle, WA 98103 Cell division depends critically on the temporally con- In eukaryotic cells, duplicated chromosomes must be trolled assembly of mitotic spindles, which are responsible symmetrically partitioned to opposite ends of the cell by the for the distribution of duplicated chromosomes to each activities of the mitotic spindle. During mitosis, spindles are as- of the two daughter cells. To gain insight into the pro- sembled, chromosomes are partitioned, and the spindles are then cess, Vizeacoumar et al., in this issue (Vizeacoumar et al. disassembled. The fidelity of this process is critical to ensure equal 2010. J. Cell Biol. doi:10.1083/jcb.200909013), have chromosome segregation during division and maintenance of combined systems genetics with high-throughput and proper chromosome number. In higher eukaryotes, structures high-content imaging to comprehensively identify and called centrosomes serve as central organizers of the mitotic classify novel components that contribute to the morphol- spindle. In yeast, spindle pole bodies are structurally distinct ogy and function of the mitotic spindle. from centrosomes, but perform an analogous function. At the start of the cell cycle, cells have a single spindle pole body embed- ded in the nuclear envelope. The spindle pole body is duplicated When, in the mid-1930s, Professor Øjvind Winge at the Carls- early in the cell cycle, and microtubules associate with and radi- berg Laboratory in Denmark discovered the sexual practices of ate from the structure (Byers and Goetsch, 1975). As the cell brewer’s yeast (Winge, 1935), he set in motion an era of scientists cycle progresses, the microtubules associate with the cortices exploiting Saccharomyces cerevisiae as an experimental model of the mother and the budding daughter cell, pulling one of the system for biological research. -

Mitochondrial Diseases: Expanding the Diagnosis in the Era of Genetic Testing
Saneto. J Transl Genet Genom 2020;4:384-428 Journal of Translational DOI: 10.20517/jtgg.2020.40 Genetics and Genomics Review Open Access Mitochondrial diseases: expanding the diagnosis in the era of genetic testing Russell P. Saneto1,2 1Center for Integrative Brain Research, Neuroscience Institute, Seattle, WA 98101, USA. 2Department of Neurology/Division of Pediatric Neurology, Seattle Children’s Hospital/University of Washington, Seattle, WA 98105, USA. Correspondence to: Dr. Russell P. Saneto, Department of Neurology/Division of Pediatric Neurology, Seattle Children’s Hospital/ University of Washington, 4800 Sand Point Way NE, Seattle, WA 98105, USA. E-mail: [email protected] How to cite this article: Saneto RP. Mitochondrial diseases: expanding the diagnosis in the era of genetic testing. J Transl Genet Genom 2020;4:348-428. http://dx.doi.org/10.20517/jtgg.2020.40 Received: 29 Jun 2020 First Decision: 27 Jul 2020 Revised: 15 Aug 2020 Accepted: 21 Aug 2020 Available online: 29 Sep 2020 Academic Editor: Andrea L. Gropman Copy Editor: Cai-Hong Wang Production Editor: Jing Yu Abstract Mitochondrial diseases are clinically and genetically heterogeneous. These diseases were initially described a little over three decades ago. Limited diagnostic tools created disease descriptions based on clinical, biochemical analytes, neuroimaging, and muscle biopsy findings. This diagnostic mechanism continued to evolve detection of inherited oxidative phosphorylation disorders and expanded discovery of mitochondrial physiology over the next two decades. Limited genetic testing hampered the definitive diagnostic identification and breadth of diseases. Over the last decade, the development and incorporation of massive parallel sequencing has identified approximately 300 genes involved in mitochondrial disease. -
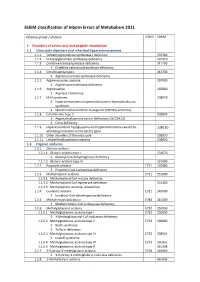
SSIEM Classification of Inborn Errors of Metabolism 2011
SSIEM classification of Inborn Errors of Metabolism 2011 Disease group / disease ICD10 OMIM 1. Disorders of amino acid and peptide metabolism 1.1. Urea cycle disorders and inherited hyperammonaemias 1.1.1. Carbamoylphosphate synthetase I deficiency 237300 1.1.2. N-Acetylglutamate synthetase deficiency 237310 1.1.3. Ornithine transcarbamylase deficiency 311250 S Ornithine carbamoyltransferase deficiency 1.1.4. Citrullinaemia type1 215700 S Argininosuccinate synthetase deficiency 1.1.5. Argininosuccinic aciduria 207900 S Argininosuccinate lyase deficiency 1.1.6. Argininaemia 207800 S Arginase I deficiency 1.1.7. HHH syndrome 238970 S Hyperammonaemia-hyperornithinaemia-homocitrullinuria syndrome S Mitochondrial ornithine transporter (ORNT1) deficiency 1.1.8. Citrullinemia Type 2 603859 S Aspartate glutamate carrier deficiency ( SLC25A13) S Citrin deficiency 1.1.9. Hyperinsulinemic hypoglycemia and hyperammonemia caused by 138130 activating mutations in the GLUD1 gene 1.1.10. Other disorders of the urea cycle 238970 1.1.11. Unspecified hyperammonaemia 238970 1.2. Organic acidurias 1.2.1. Glutaric aciduria 1.2.1.1. Glutaric aciduria type I 231670 S Glutaryl-CoA dehydrogenase deficiency 1.2.1.2. Glutaric aciduria type III 231690 1.2.2. Propionic aciduria E711 232000 S Propionyl-CoA-Carboxylase deficiency 1.2.3. Methylmalonic aciduria E711 251000 1.2.3.1. Methylmalonyl-CoA mutase deficiency 1.2.3.2. Methylmalonyl-CoA epimerase deficiency 251120 1.2.3.3. Methylmalonic aciduria, unspecified 1.2.4. Isovaleric aciduria E711 243500 S Isovaleryl-CoA dehydrogenase deficiency 1.2.5. Methylcrotonylglycinuria E744 210200 S Methylcrotonyl-CoA carboxylase deficiency 1.2.6. Methylglutaconic aciduria E712 250950 1.2.6.1. Methylglutaconic aciduria type I E712 250950 S 3-Methylglutaconyl-CoA hydratase deficiency 1.2.6.2. -

Carrier Screening Panel
Advancing Non-Invasive Healthcare Cell3TM Target: Carrier Screening Panel Detects 448 childhood recessive disorders DISORDER GENE DISEASE TYPE 2-Methylbutryl-CoA dehydrogenase deficiency ACADSB Metabolic 3-Hydroxy-3-methylglutaryl- CoA lyase deficiency HMGCL Metabolic 3-Hydroxyacyl- CoA dehydrogenase deficiency HADH Metabolic 3-Methylcrotonyl- CoA carboxylase 2 deficiency MCCC2 Metabolic 3-Methylglutaconic aciduria, i AUH Metabolic 3-Methylglutaconic aciduria, iii OPA3 Metabolic 3-Methylglutaconic aciduria, v DNAJC19 Metabolic α -Thalassemia/mental retardation syndrome, nondeletion, XLR ATRX ATRX Hematologic ABCD syndrome EDNRB Cutaneous Abetalipoproteinemia; ABL MTTP Metabolic Achalasia-Addisonianism-Alacrima syndrome; AAA AAAS Endocrine Achondrogenesis, Ib; ACG1b SLC26A2 Skeletal Achromatopsia 3; ACHM3 CNGB3 Ocular Acrocallosal syndrome; ACLS GLI3 Developmental Acyl-CoA dehydrogenase family, member 9, deficiency of ACAD9 Metabolic Acyl-CoA dehydrogenase, long-chain, deficiency of ACADL Metabolic Acyl-CoA dehydrogenase, medium-chain, deficiency of ACADM Metabolic Acyl-CoA dehydrogenase, short-chain, deficiency of ACADS Metabolic Acyl-CoA dehydrogenase, very long-chain, deficiency of ACADVL Metabolic Adrenal hyperplasia, congenital, due to 21-hydroxylase deficiency CYP21A2 Endocrine Adrenoleukodystrophy; ALD ABCD1 Neurological Afibrinogenemia, congenital FGA Hematologic Afibrinogenemia, congenital FGB Hematologic Afibrinogenemia, congenital FGG Hematologic Agammaglobulinemia, XLR, XLA BTK Immunodeficiency Agenesis of the corpus callosum