A Mouse Translocation Associated with Caspr5-2 Disruption and Perinatal Lethality
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Rabbit Anti-CASPR/FITC Conjugated Antibody-SL11128R-FITC
SunLong Biotech Co.,LTD Tel: 0086-571- 56623320 Fax:0086-571- 56623318 E-mail:[email protected] www.sunlongbiotech.com Rabbit Anti-CASPR/FITC Conjugated antibody SL11128R-FITC Product Name: Anti-CASPR/FITC Chinese Name: FITC标记的轴突蛋白4/少突胶质细胞抗体 Neurexin4; caspr 1; Caspr; Caspr1; Cntnap 1; Cntnap1; CNTP 1; CNTP1; CNTP1_HUMAN; Contactin associated protein 1; Contactin-associated protein 1; Alias: MHDNIV; NCP 1; NCP1; Neurexin 4; Neurexin IV; Neurexin-4; Nrxn 4; Nrxn4; p190; Paranodin. Organism Species: Rabbit Clonality: Polyclonal React Species: Human,Mouse,Rat,Dog,Pig,Cow,Horse, Flow-Cyt=1:50-200ICC=1:50-200IF=1:50-200 Applications: not yet tested in other applications. optimal dilutions/concentrations should be determined by the end user. Molecular weight: 154kDa Cellular localization: The cell membrane Form: Lyophilized or Liquid Concentration: 1mg/ml immunogen: KLH conjugated synthetic peptide derived from human CASPR/Neurexin4 Lsotype: IgGwww.sunlongbiotech.com Purification: affinity purified by Protein A Storage Buffer: 0.01M TBS(pH7.4) with 1% BSA, 0.03% Proclin300 and 50% Glycerol. Store at -20 °C for one year. Avoid repeated freeze/thaw cycles. The lyophilized antibody is stable at room temperature for at least one month and for greater than a year Storage: when kept at -20°C. When reconstituted in sterile pH 7.4 0.01M PBS or diluent of antibody the antibody is stable for at least two weeks at 2-4 °C. background: Neurexins comprise a family of neuronal cell surface proteins, which include neurexin I (NRXN1), neurexin II (NRXN2), neurexin III (NRXN3) and CASPR (neurexin IV). Product Detail: Neurexins I-III are expressed as ? and ∫ isoforms. -

(Caspr) and Contactin Form a Complex That Is Targeted to the Paranodal Junctions During Myelination
The Journal of Neuroscience, November 15, 2000, 20(22):8354–8364 Contactin-Associated Protein (Caspr) and Contactin Form a Complex That Is Targeted to the Paranodal Junctions during Myelination Jose C. Rios,1 Carmen V. Melendez-Vasquez,1 Steven Einheber,1 Marc Lustig,2 Martin Grumet,2 John Hemperly,5 Elior Peles,6 and James L. Salzer1,3,4 Departments of 1Cell Biology, 2Pharmacology, 3Neurology, and the 4Kaplan Cancer Center, New York University School of Medicine, New York, New York 10016, 5BD Technologies, Research Triangle Park, North Carolina 27709, and 6Department of Molecular Cell Biology, The Weizmann Institute of Science, Rehovot 76100, Israel Specialized paranodal junctions form between the axon and the associated specifically with Caspr in the paranodes, whereas a closely apposed paranodal loops of myelinating glia. They are higher-molecular-weight form of contactin, not associated with interposed between sodium channels at the nodes of Ranvier Caspr, is present in central nodes of Ranvier. These results and potassium channels in the juxtaparanodal regions; their suggest that the targeting of contactin to different axonal do- precise function and molecular composition have been elusive. mains may be determined, in part, via its association with Caspr. We previously reported that Caspr (contactin-associated protein) Treatment of myelinating cocultures of Schwann cells and neu- is a major axonal constituent of these junctions (Einheber et al., rons with RPTP–Fc, a soluble construct containing the carbonic 1997). We now report that contactin colocalizes and forms a cis anhydrase domain of the receptor protein tyrosine phosphatase complex with Caspr in the paranodes and juxtamesaxon. -

LRP1 Regulates Peroxisome Biogenesis and Cholesterol
RESEARCH ARTICLE LRP1 regulates peroxisome biogenesis and cholesterol homeostasis in oligodendrocytes and is required for proper CNS myelin development and repair Jing-Ping Lin1, Yevgeniya A Mironova2, Peter Shrager3, Roman J Giger1,2,4,5* 1Department of Cell and Developmental Biology, University of Michigan School of Medicine, Ann Arbor, MI, United States; 2Cellular and Molecular Biology Graduate Program, University of Michigan Medical School, Ann Arbor, MI, United States; 3Department of Neuroscience, University of Rochester Medical Center, Rochester, NY, United States ; 4Department of Neurology, University of Michigan Medical School, Ann Arbor, MI, United States; 5Interdepartmental Neuroscience Graduate Program, University of Michigan Medical School, Ann Arbor, MI, United States Abstract Low-density lipoprotein receptor-related protein-1 (LRP1) is a large endocytic and signaling molecule broadly expressed by neurons and glia. In adult mice, global inducible (Lrp1flox/ flox;CAG-CreER) or oligodendrocyte (OL)-lineage specific ablation (Lrp1flox/flox;Pdgfra-CreER) of Lrp1 attenuates repair of damaged white matter. In oligodendrocyte progenitor cells (OPCs), Lrp1 is required for cholesterol homeostasis and differentiation into mature OLs. Lrp1-deficient OPC/ OLs show a strong increase in the sterol-regulatory element-binding protein-2 yet are unable to maintain normal cholesterol levels, suggesting more global metabolic deficits. Mechanistic studies revealed a decrease in peroxisomal biogenesis factor-2 and fewer peroxisomes in OL processes. / *For correspondence: Treatment of Lrp1À À OPCs with cholesterol or activation of peroxisome proliferator-activated [email protected] receptor-g with pioglitazone alone is not sufficient to promote differentiation; however, when combined, cholesterol and pioglitazone enhance OPC differentiation into mature OLs. Collectively, Competing interests: The our studies reveal a novel role for Lrp1 in peroxisome biogenesis, lipid homeostasis, and OPC authors declare that no competing interests exist. -

Anti-CASPR (MOUSE) Monoclonal Antibody - 200-301-F03
Anti-CASPR (MOUSE) Monoclonal Antibody - 200-301-F03 Code: 200-301-F03 Size: 100 µg Product Description: Anti-CASPR (MOUSE) Monoclonal Antibody - 200-301-F03 Concentration: 1mg/mL by UV absorbance at 280 nm PhysicalState: Liquid (sterile filtered) Label Unconjugated Host Mouse Gene Name CNTNAP1 Species Reactivity Human, Mouse, Rat Buffer 0.02 M Potassium Phosphate, 0.15 M Sodium Chloride, pH 7.2 Stabilizer 50% (v/v) Glycerol Storage Condition Store vial at -20° C prior to opening. Aliquot contents and freeze at -20° C or below for extended storage. Avoid cycles of freezing and thawing. Centrifuge product if not completely clear after standing at room temperature. This product is stable for several weeks at 4° C as an undiluted liquid. Dilute only prior to immediate use. Synonyms CASPR1, CNTP1, Contactin associated protein 1, NCP1, Paranodin, Neurexin 4, p190, CASPR, NRXN4 Application Note Anti-CASPR Antibody is suitable for use in WB, IP, and IHC. Expect a band approximately ~220kDa on specific lysates. Specific conditions for reactivity should be optimized by the end user. Background Caspr (contactin-associated protein) is a part of the neurexin family. It lies in the paranodal section of the myelin sheath. It’s role is for myelin sheath attachment along with contactin in a cis-complex. Caspr and Caspr2 regulate the formation of distinct axonal domains around the nodes of Ranvier. Caspr is required for the generation of a membrane barrier at the paranodal junction, whereas Caspr serves as a membrane scaffold that clusters Kv1 channels at the juxtaparanodal region. Both interact with protein 4.1B. -

Whole-Exome Sequencing Supports Genetic Heterogeneity in Childhood
Worthey et al. Journal of Neurodevelopmental Disorders 2013, 5:29 http://www.jneurodevdisorders.com/content/5/1/29 RESEARCH Open Access Whole-exome sequencing supports genetic heterogeneity in childhood apraxia of speech Elizabeth A Worthey1,2, Gordana Raca3, Jennifer J Laffin4, Brandon M Wilk2, Jeremy M Harris2, Kathy J Jakielski5, David P Dimmock1,2, Edythe A Strand6 and Lawrence D Shriberg7* Abstract Background: Childhood apraxia of speech (CAS) is a rare, severe, persistent pediatric motor speech disorder with associated deficits in sensorimotor, cognitive, language, learning and affective processes. Among other neurogenetic origins, CAS is the disorder segregating with a mutation in FOXP2 in a widely studied, multigenerational London family. We report the first whole-exome sequencing (WES) findings from a cohort of 10 unrelated participants, ages 3 to 19 years, with well-characterized CAS. Methods: As part of a larger study of children and youth with motor speech sound disorders, 32 participants were classified as positive for CAS on the basis of a behavioral classification marker using auditory-perceptual and acoustic methods that quantify the competence, precision and stability of a speaker’s speech, prosody and voice. WES of 10 randomly selected participants was completed using the Illumina Genome Analyzer IIx Sequencing System. Image analysis, base calling, demultiplexing, read mapping, and variant calling were performed using Illumina software. Software developed in-house was used for variant annotation, prioritization and interpretation to identify those variants likely to be deleterious to neurodevelopmental substrates of speech-language development. Results: Among potentially deleterious variants, clinically reportable findings of interest occurred on a total of five chromosomes (Chr3, Chr6, Chr7, Chr9 and Chr17), which included six genes either strongly associated with CAS (FOXP1 and CNTNAP2) or associated with disorders with phenotypes overlapping CAS (ATP13A4, CNTNAP1, KIAA0319 and SETX). -

Localization of Caspr2 in Myelinated Nerves Depends on Axon–Glia Interactions and the Generation of Barriers Along the Axon
The Journal of Neuroscience, October 1, 2001, 21(19):7568–7575 Localization of Caspr2 in Myelinated Nerves Depends on Axon–Glia Interactions and the Generation of Barriers along the Axon Sebastian Poliak,1 Leora Gollan,1 Daniela Salomon,1 Erik O. Berglund,2 Reiko Ohara,3 Barbara Ranscht,2 and Elior Peles1 1Department of Molecular Cell Biology, The Weizmann Institute of Science, Rehovot 76100, Israel, 2The Burnham Institute, Neurobiology Program, La Jolla, California 92037, and 3Department of Human Gene Research at Kazusa DNA Research Institute, Chiba 292-0812, Japan Cell recognition proteins of the contactin-associated protein before relocating to the adjacent juxtaparanodal region. This (Caspr) family demarcate distinct domains along myelinated transition was not observed in CGT mice, where Caspr2 and axons. Caspr is present at the paranodal junction formed be- Kv1.2 remained paranodal. Double labeling for Caspr and tween the axon and myelinating glial cells, whereas Caspr2 is Caspr2 demonstrated that these two related proteins occupied localized and associates with K ϩ channels at the adjacent mutually excluding domains along the axon and revealed the juxtaparanodal region. Here we investigated the distribution of presence of both paranodal and internodal barrier-like struc- Caspr2 during development of peripheral nerves of normal and tures that are delineated by Caspr. Finally, we found that the galactolipids-deficient [ceramide galactosyl transferase disruption of axon–glia contact in CGTϪ/Ϫ nerves also affects (CGT)Ϫ/Ϫ] mice. This mutant exhibits paranodal abnormalities, the localization of the cytoskeleton-associated protein 4.1B lacking all putative adhesion components of this junction, in- along the axon. -

Conservation of Core Gene Expression in Vertebrate Tissues
Open Access Research article Conservation of core gene expression in vertebrate tissues Esther T Chan*¶, Gerald T Quon†¶, Gordon Chua‡¶¥, Tomas Babak¶#, Miles Trochesset†‡, Ralph A Zirngibl*, Jane Aubin*, Michael JH Ratcliffe§, Andrew Wilde*, Michael Brudno†‡¶, Quaid D Morris*†‡¶ and Timothy R Hughes*‡¶ Addresses: *Department of Molecular Genetics, †Department of Computer Science, ‡Banting and Best Department of Medical Research, §Department of Immunology and Sunnybrook Research Institute, and ¶Terrence Donnelly Centre for Cellular and Biomolecular Research, University of Toronto, 160 College Street, Toronto, Ontario M5S 3E1, Canada. ¥Current address: Department of Biological Sciences, University of Calgary, 2500 University Drive NW, Calgary, Alberta, T2N 1N4 Canada. #Current address: Rosetta Inpharmatics, 401 Terry Avenue North, Seattle, WA 98109, USA. Correspondence: Quaid D Morris. Email: [email protected]. Timothy R Hughes. Email: [email protected] Published: 16 April 2009 Received: 23 January 2009 Revised: 12 March 2009 Journal of Biology 2009, 8:33 (doi:10.1186/jbiol130) Accepted: 18 March 2009 The electronic version of this article is the complete one and can be found online at http://jbiol.com/content/8/3/33 © 2009 Chan et al.; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background Vertebrates share the same general body plan and organs, possess related sets of genes, and rely on similar physiological mechanisms, yet show great diversity in morphology, habitat and behavior. -

Vestibular Damage and Repair in Chronic Ototoxicity: Cellular Stages, Physiological Deficits and Molecular Mechanisms
Vestibular Damage and Repair in Chronic Ototoxicity: Cellular Stages, Physiological Deficits and Molecular Mechanisms Erin A. Greguske ADVERTIMENT. La consulta d’aquesta tesi queda condicionada a l’acceptació de les següents condicions d'ús: La difusió d’aquesta tesi per mitjà del servei TDX (www.tdx.cat) i a través del Dipòsit Digital de la UB (diposit.ub.edu) ha estat autoritzada pels titulars dels drets de propietat intelꞏlectual únicament per a usos privats emmarcats en activitats d’investigació i docència. No s’autoritza la seva reproducció amb finalitats de lucre ni la seva difusió i posada a disposició des d’un lloc aliè al servei TDX ni al Dipòsit Digital de la UB. No s’autoritza la presentació del seu contingut en una finestra o marc aliè a TDX o al Dipòsit Digital de la UB (framing). Aquesta reserva de drets afecta tant al resum de presentació de la tesi com als seus continguts. En la utilització o cita de parts de la tesi és obligat indicar el nom de la persona autora. ADVERTENCIA. La consulta de esta tesis queda condicionada a la aceptación de las siguientes condiciones de uso: La difusión de esta tesis por medio del servicio TDR (www.tdx.cat) y a través del Repositorio Digital de la UB (diposit.ub.edu) ha sido autorizada por los titulares de los derechos de propiedad intelectual únicamente para usos privados enmarcados en actividades de investigación y docencia. No se autoriza su reproducción con finalidades de lucro ni su difusión y puesta a disposición desde un sitio ajeno al servicio TDR o al Repositorio Digital de la UB. -

C-Fibers May Modulate Adjacent Aδ-Fibers Through Axon-Axon CGRP Signaling at Nodes of Ranvier in the Trigeminal System Jacob C
Edvinsson et al. The Journal of Headache and Pain (2019) 20:105 The Journal of Headache https://doi.org/10.1186/s10194-019-1055-3 and Pain RESEARCH ARTICLE Open Access C-fibers may modulate adjacent Aδ-fibers through axon-axon CGRP signaling at nodes of Ranvier in the trigeminal system Jacob C. A. Edvinsson1,4, Karin Warfvinge1,2, Diana N. Krause2,3, Frank W. Blixt5, Majid Sheykhzade4, Lars Edvinsson1,2* and Kristian A. Haanes1 Abstract Background: Monoclonal antibodies (mAbs) towards CGRP or the CGRP receptor show good prophylactic antimigraine efficacy. However, their site of action is still elusive. Due to lack of passage of mAbs across the blood- brain barrier the trigeminal system has been suggested a possible site of action because it lacks blood-brain barrier and hence is available to circulating molecules. The trigeminal ganglion (TG) harbors two types of neurons; half of which store CGRP and the rest that express CGRP receptor elements (CLR/RAMP1). Methods: With specific immunohistochemistry methods, we demonstrated the localization of CGRP, CLR, RAMP1, and their locations related to expression of the paranodal marker contactin-associated protein 1 (CASPR). Furthermore, we studied functional CGRP release separately from the neuron soma and the part with only nerve fibers of the trigeminal ganglion, using an enzyme-linked immunosorbent assay. Results: Antibodies towards CGRP and CLR/RAMP1 bind to two different populations of neurons in the TG and are found in the C- and the myelinated Aδ-fibers, respectively, within the dura mater and in trigeminal ganglion (TG). CASPR staining revealed paranodal areas of the different myelinated fibers inhabiting the TG and dura mater. -
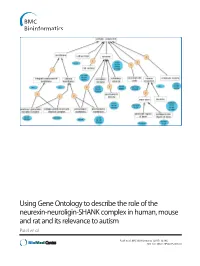
Using Gene Ontology to Describe the Role of the Neurexin-Neuroligin-SHANK Complex in Human, Mouse and Rat and Its Relevance to Autism Patel Et Al
Using Gene Ontology to describe the role of the neurexin-neuroligin-SHANK complex in human, mouse and rat and its relevance to autism Patel et al. Patel et al. BMC Bioinformatics (2015) 16:186 DOI 10.1186/s12859-015-0622-0 Patel et al. BMC Bioinformatics (2015) 16:186 DOI 10.1186/s12859-015-0622-0 METHODOLOGY ARTICLE Open Access Using Gene Ontology to describe the role of the neurexin-neuroligin-SHANK complex in human, mouse and rat and its relevance to autism Sejal Patel1,2,3, Paola Roncaglia4,5 and Ruth C. Lovering3* Abstract Background: People with an autistic spectrum disorder (ASD) display a variety of characteristic behavioral traits, including impaired social interaction, communication difficulties and repetitive behavior. This complex neurodevelopment disorder is known to be associated with a combination of genetic and environmental factors. Neurexins and neuroligins play a key role in synaptogenesis and neurexin-neuroligin adhesion is one of several processes that have been implicated in autism spectrum disorders. Results: In this report we describe the manual annotation of a selection of gene products known to be associated with autism and/or the neurexin-neuroligin-SHANK complex and demonstrate how a focused annotation approach leads to the creation of more descriptive Gene Ontology (GO) terms, as well as an increase in both the number of gene product annotations and their granularity, thus improving the data available in the GO database. Conclusions: The manual annotations we describe will impact on the functional analysis of a variety of future autism-relevant datasets. Comprehensive gene annotation is an essential aspect of genomic and proteomic studies, as the quality of gene annotations incorporated into statistical analysis tools affects the effective interpretation of data obtained throughgenomewideassociationstudies, next generation sequencing, proteomic and transcriptomic datasets. -

Caspr3 and Caspr4, Two Novel Members of the Caspr Family Are Expressed in the Nervous System and Interact with PDZ Domains
Molecular and Cellular Neuroscience 20, 283–297 (2002) doi:10.1006/mcne.2002.1110 MCN Caspr3 and Caspr4, Two Novel Members of the Caspr Family Are Expressed in the Nervous System and Interact with PDZ Domains Ivo Spiegel,* Daniela Salomon,* Beat Erne,† Nicole Schaeren-Wiemers,† and Elior Peles* *Department of Molecular Cell Biology, The Weizmann Institute of Science, Rehovot 76100, Israel; and †Neurobiology Laboratory, Department of Research, University Hospital Basel, Pharmacenter, CH-4056 Basel, Switzerland The NCP family of cell-recognition molecules represents a sive alternative splicing to generate a large number of distinct subgroup of the neurexins that includes Caspr and isoforms that are differentially expressed throughout the Caspr2, as well as Drosophila Neurexin-IV and axotactin. brain (Ullrich et al., 1995; Puschel and Betz, 1995). The Here, we report the identification of Caspr3 and Caspr4, two extracellular region of the neurexins binds to several new NCPs expressed in nervous system. Caspr3 was de- tected along axons in the corpus callosum, spinal cord, membrane bound ligands, including neuroligins (Icht- basket cells in the cerebellum and in peripheral nerves, as chenko et al., 1995; Nguyen and Sudhof, 1997; Scheiffele et well as in oligodendrocytes. In contrast, expression of al., 2000) and dystroglycan (Sugita et al., 2001), suggesting Caspr4 was more restricted to specific neuronal subpopu- their involvement in cell–cell interactions and synapse lations in the olfactory bulb, hippocampus, deep cerebellar formation. nuclei, and the substantia nigra. Similar to the neurexins, the The NCP family (for Neurexin-IV/Caspr/Paranodin; cytoplasmic tails of Caspr3 and Caspr4 interacted differen- Bellen et al., 1998) represents a distinct subgroup of the tially with PDZ domain-containing proteins of the CASK/ neurexins that includes mammalian Caspr (Contactin Lin2-Veli/Lin7-Mint1/Lin10 complex. -

A Novel Caspr Mutation Causes the Shambling Mouse Phenotype by Disrupting Axoglial Interactions of Myelinated Nerves
J Neuropathol Exp Neurol Vol. 68, No. 11 Copyright Ó 2009 by the American Association of Neuropathologists, Inc. November 2009 pp. 1207Y1218 ORIGINAL ARTICLE A Novel Caspr Mutation Causes the Shambling Mouse Phenotype by Disrupting Axoglial Interactions of Myelinated Nerves Xiao-yang Sun, PhD, Yoshiko Takagishi, PhD, Erina Okabe, MSc, Yuˆko Chishima, BSc, Yasuhiko Kanou, PhD, Shiori Murase, DVM, Kazue Mizumura, MD, PhD, Mie Inaba, MD, Yukio Komatsu, MD, PhD, Yoshitaka Hayashi, MD, PhD, Elior Peles, PhD, Sen-ichi Oda, PhD, and Yoshiharu Murata, MD, PhD the node of Ranvier, the paranode, the juxtaparanode, and Abstract the internode. The paranodes flanking the node of Ranvier The neurological mouse mutation shambling (shm) exhibits ataxia are one of the major attachment sites between the axon and and hindlimb paresis. Positional cloning of shm showed that it en- myelinating glial cells, that is, oligodendrocytes in the central codes contactin-associated protein (Caspr), which is required for nervous system (CNS) and Schwann cells in the peripheral formation of the paranodal junction in myelinated nerves. The shm nervous system (PNS) (1Y5). A septate-like adhesive junction mutation is a TT insertion in the Caspr gene that results in a frame is formed at the paranode between the axolemma and myelin shift and a premature stop codon at the COOH-terminus. The trun- loops. The paranode acts as an electrical and a biochemical cated Caspr protein that is generated lacks the transmembrane and barrier between nodal and internodal membrane compart- cytoplasmic domains. Here, we found that the nodal/paranodal ments. In addition, the paranodal junction is thought to play a axoplasm of shm mice lack paranodal junctions and contain large role in molecular communication between the neuron and the mitochondria and abnormal accumulations of cytoplasmic organ- glial cell.