Specific PCR Test for the Quick Recognition of Equine Tissue in Raw
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Smoking the Perfect Sausage
PROCEDURES FOR SMOKING THE PERFECT SAUSAGE Start with a stuffed casing at room temperature. DRYING THE SAUSAGE You can achieve the drying by placing the sausage in your smokehouse with the damper open at about 140-150° for one hour. Reasons for drying the sausage: Drying the sausage brings all the sausages to about the same temperature for an even smoke color. Drying conditions the surface of the sausage to ready it to accept smoke. Drying causes a “skin” to form on the outside surface of the sausage. Drying gives the collagen casing strength to hold up during cooking. Drying also attaches the casing to the sausage so as to avoid forming a fat layer between the sausage and the casing. SMOKING THE SAUSAGE Smoking can be achieved by placing a pan of sawdust/chips in the smoker on the burner. The sawdust/chips must be soaked in water at least one hour. Soak in half the volume of water that you have sawdust/chips. (4 cups sawdust/2 cups water) Heat the smoker to approximately 170⁰ to ignite the sawdust/chips to achieve smoke. Close the damper to half open at this point. COOKING As the sawdust/chips burn, the water will evaporate and a dry heat will set in. The dry smoke will set the smoke in the sausage. After most of the sawdust/chips have burned, remove the pan from the smoker and let the pan cool for 5-10 minutes. After this time, fill the pan half full of water and return to the burner. Close the damper and turn the temperature to approximately 180-190⁰, this will cause a high humidity to cook the sausage. -

The Evaluation of Pathogen Survival in Dry Cured Charcuterie Style Sausages
University of Kentucky UKnowledge Theses and Dissertations--Animal and Food Sciences Animal and Food Sciences 2019 THE EVALUATION OF PATHOGEN SURVIVAL IN DRY CURED CHARCUTERIE STYLE SAUSAGES Jennifer Michelle McNeil University of Kentucky, [email protected] Digital Object Identifier: https://doi.org/10.13023/etd.2019.074 Right click to open a feedback form in a new tab to let us know how this document benefits ou.y Recommended Citation McNeil, Jennifer Michelle, "THE EVALUATION OF PATHOGEN SURVIVAL IN DRY CURED CHARCUTERIE STYLE SAUSAGES" (2019). Theses and Dissertations--Animal and Food Sciences. 102. https://uknowledge.uky.edu/animalsci_etds/102 This Master's Thesis is brought to you for free and open access by the Animal and Food Sciences at UKnowledge. It has been accepted for inclusion in Theses and Dissertations--Animal and Food Sciences by an authorized administrator of UKnowledge. For more information, please contact [email protected]. STUDENT AGREEMENT: I represent that my thesis or dissertation and abstract are my original work. Proper attribution has been given to all outside sources. I understand that I am solely responsible for obtaining any needed copyright permissions. I have obtained needed written permission statement(s) from the owner(s) of each third-party copyrighted matter to be included in my work, allowing electronic distribution (if such use is not permitted by the fair use doctrine) which will be submitted to UKnowledge as Additional File. I hereby grant to The University of Kentucky and its agents the irrevocable, non-exclusive, and royalty-free license to archive and make accessible my work in whole or in part in all forms of media, now or hereafter known. -

Nutritional Characteristics and Consumer Acceptability of Sausages with Different Combinations of Goat and Beef Meats
Functional Foods in Health and Disease 2016; 6(1):42-58 Page 42 of 58 Research Article Open Access Nutritional characteristics and consumer acceptability of sausages with different combinations of goat and beef meats *Fatemeh Malekian, Margarita Khachaturyan, Sebhatu Gebrelul, and James F. Henson Southern University Agricultural Research and Extension Center, A. O. Williams Hall, P. O. Box 10010, Baton Rouge, Louisiana 70813, USA *Corresponding author: Fatemeh Malekian, Professor, Southern University Agricultural Research and Extension Center, 181 B.A. Little Drive, A.O. Williams Hall, P.O. Box 10010, Baton Rouge LA 70813, USA Submission date: October 28, 2015, Acceptance date: January 28, 2016: Publication date: January 29, 2016 ABSTRACT Background: Obesity and cardiovascular heart diseases are becoming more prevalent in the United States. This is partly due to the increasing consumption of red meats, such as pork and beef. However, goat meat has the potential to replace these traditionally consumed meats. Rice bran is a rich source of antioxidants, including vitamin E, which can be utilized as a binder in meat and meat-related products. Methods: Goat meat/beef sausages were formulated to contain either 50/50, 75/25 or 100/0 percent goat meat/beef, with either no added rice bran (NRB) or 3 percent stabilized rice bran (RB). Proximate analysis, fatty acids, 훼-tocopherol and cholesterol concentrations of the six cooked formulations were determined. The six sausage formulations were compared in a consumer acceptability taste test. Results: The fat concentration of the NRB and RB formulations decreased linearly with increasing percentages of goat meat (p < 0.001). -

Microbial Characterization of Horse Meat Dry Sausage
Microbial characterization of horse meat dry sausage Microbial characterization of horse meat dry sausage ;/"/457&/04536ė/*%*0 Microbial characterization Table 1 Methodology of microbiological analyses per, garlic, nitrite salt). After filling Parameter Growth medium Incubation in the natural casing and draining Aerobic mesophilic bacteria Plate Count Agar (PCA) $IPVST on the sticks, cold smoking followed of horse meat dry sausage De Man Ragosa Sharpe for 3 days and then ripening in the Lactic acid bacteria $IPVST agar (MRS) fermentation chamber for 33 days. Coagulase-negative cocci Manitol Salt Agar (MSA) $IPVST Sampling was done every time with S. aureus Baird-Parker agar (BP) $IPVST three sausages sampled on the day Alagić, D. 1, N. Zdolec 2, B. Njari 2, I. Filipović 2, A. Ekert-Kabalin 3, G. Ćorić-Alagić 4, M. Stojnović 1, N. Vragović 5, L. Kozačinski 2 Violet red bile glucose BOE"UUIFCFHJOOJOHPG Enterobacteria $IPVST agar (VRBG) production process sampling was scientific paper E. coli Coli ID $IPVST done also for the raw material (horse Oxytetracycline yeast agar Yeasts and moulds $EBZT meat and pork fat tissue). The sam- with tetracycline (OGY) Kanamycin esculin agar ples were transported to the labora- Enterococci $IPVST (KEA) UPSZJOBQPSUBCMFSFGSJHFSBUPS C). Cetrimide-fucidin- Pseudomonas spp. $IPVST All samples were analyzed for micro- Summary cephaloridine ( CFC) agar The aim of this study was to evaluate microbiological changes of traditionally produced horse meat sausages depending on maturing Sulphyte Polymixine biological analyses in triplicate. Sulphite reducing clostridia $EBZT phases and season, to determine lactic acid bacteria and to research their inhibitory potential towards L. monocytogenes. Produc- Suphadiazine agar (SPS) Buffered peptonic water tion season influenced significantly on total viable count, lactic acid bacteria, coagulase-negative cocci, enterococci and yeasts in $IPVST Microbiological analyses Rappaport-Vasiliadis broth final product (p<0.05). -

We're A' Jock Tamson's Bairns
Some hae meat and canna eat, BIG DISHES SIDES Selkirk and some wad eat that want it, Frying Scotsman burger £12.95 Poke o' ChipS £3.45 Buffalo Farm beef burger, haggis fritter, onion rings. whisky cream sauce & but we hae meat and we can eat, chunky chips Grace and sae the Lord be thankit. Onion Girders & Irn Bru Mayo £2.95 But 'n' Ben Burger £12.45 Buffalo Farm beef burger, Isle of Mull cheddar, lettuce & tomato & chunky chips Roasted Roots £2.95 Moving Munros (v)(vg) £12.95 Hoose Salad £3.45 Wee Plates Mooless vegan burger, vegan haggis fritter, tomato chutney, pickles, vegan cheese, vegan bun & chunky chips Mashed Tatties £2.95 Soup of the day (v) £4.45 Oor Famous Steak Pie £13.95 Served piping hot with fresh baked sourdough bread & butter Steak braised long and slow, encased in hand rolled golden pastry served with Champit Tatties £2.95 roasted roots & chunky chips or mash Cullen skink £8.95 Baked Beans £2.95 Traditional North East smoked haddock & tattie soup, served in its own bread Clan Mac £11.95 bowl Macaroni & three cheese sauce with Isle of Mull, Arron smoked cheddar & Fresh Baked Sourdough Bread & Butter £2.95 Parmesan served with garlic sourdough bread Haggis Tower £4.95 £13.95 FREEDOM FRIES £6.95 Haggis, neeps and tatties with a whisky sauce Piper's Fish Supper Haggis crumbs, whisky sauce, fried crispy onions & crispy bacon bits Battered Peterhead haddock with chunky chips, chippy sauce & pickled onion Trio of Scottishness £5.95 £4.95 Haggis, Stornoway black pudding & white pudding, breaded baws, served with Sausage & Mash -

CHAPTER-2 Charcutierie Introduction: Charcuterie (From Either the French Chair Cuite = Cooked Meat, Or the French Cuiseur De
CHAPTER-2 Charcutierie Introduction: Charcuterie (from either the French chair cuite = cooked meat, or the French cuiseur de chair = cook of meat) is the branch of cooking devoted to prepared meat products such as sausage primarily from pork. The practice goes back to ancient times and can involve the chemical preservation of meats; it is also a means of using up various meat scraps. Hams, for instance, whether smoked, air-cured, salted, or treated by chemical means, are examples of charcuterie. The French word for a person who prepares charcuterie is charcutier , and that is generally translated into English as "pork butcher." This has led to the mistaken belief that charcuterie can only involve pork. The word refers to the products, particularly (but not limited to) pork specialties such as pâtés, roulades, galantines, crépinettes, etc., which are made and sold in a delicatessen-style shop, also called a charcuterie." SAUSAGE A simple definition of sausage would be ‘the coarse or finely comminuted (Comminuted means diced, ground, chopped, emulsified or otherwise reduced to minute particles by mechanical means) meat product prepared from one or more kind of meat or meat by-products, containing various amounts of water, usually seasoned and frequently cured .’ A sausage is a food usually made from ground meat , often pork , beef or veal , along with salt, spices and other flavouring and preserving agents filed into a casing traditionally made from intestine , but sometimes synthetic. Sausage making is a traditional food preservation technique. Sausages may be preserved by curing , drying (often in association with fermentation or culturing, which can contribute to preservation), smoking or freezing. -

The Tasty World of Charcuterie
Welcome to the DISCOVER THE FLAVORS Tasty World of Charcuterie IN YOUR CHARCUTERIE BOX Charcuterie (SHär’kooduh-ree) is how the French refer to smoked, cured or cooked meats. This box of charcuterie is packed with exciting flavors you’re sure to enjoy. These traditional methods of preservation were perfected in the days before refrigeration. Our charcuterie collections contain different combinations of the products below.* Bon appétit! You’ve probably already enjoyed charcuterie; ever had bacon, salami or ham? Pork Saucisson Sec: Ready-to-eat dry-cured pork sausage, similar to salami Slice thinly and serve with black truffle butter on bread, or pizza; dice for carbonara MAKING A CHARCUTERIE PLATE Duck Saucisson Sec: Ready-to-eat dry-cured duck sausage with warming spices A charcuterie plate is probably one of the easiest, Slice thinly and serve on charcuterie board; crumble and add to fried rice and most impressive appetizers to serve. 1. On a board or platter, arrange an assortment of ready-to-eat charcuterie. Wild Boar Saucisson Sec: Ready-to-eat dry-cured wild boar sausage, robustly flavored Dice, add to pasta salad; slice thinly for cheese and charcuterie plate 2. Add a variety of complementary sides and condiments. Our recommendations - items likely in your pantry or readily available Black Truffle Butter: Ready-to-eat butter studded with pieces of black truffle at your local grocer - include: Pairs well with eggs, bread, potatoes, pasta or popcorn; add a dollop to a steak before serving; • Something acidic like cornichons/tart pickles, mustard or olives add to a charcuterie plate; pair with saucisson sec on a slice of bread • Something sweet like chutney, raw and/or dried fruits such as figs, grapes or melon Chorizo Sausage: Ready-to-eat fully-cooked heritage-breed pork sausage with a little spice • Sliced rustic bread and/or plain crackers Slice and serve at room temperature on charcuterie board; sauté or grill for paella, soups or stews A hearty red wine always pairs well with a charcuterie plate. -
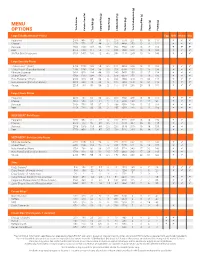
Menu Options
MENU OPTIONS Calories Total Fat Calories from (g) Fat Total (g) Fat Saturated (g) Fat Trans (mg) Cholesterol Sodium (mg) (g) Carbohydrate Total Dietary Fiber (g) (g) Sugars (g) Protein Large ExtraMostBestest® Pizzas Egg Milk Wheat Soy Pepperoni 2470 990 112 48 3.5 270 5570 251 13 14 117 a a a Cheese 2270 770 87 44 3 225 4490 253 13 12 121 a a a Sausage 2650 1130 128 53 2.5 290 5860 252 13 12 126 a a a Beef 2550 1030 117 52 5 280 5620 250 13 12 126 a a a Stuffed Crust Pepperoni 3030 1400 158 72 4.5 395 7170 259 13 15 146 a a a Large Specialty Pizzas 5 Meat Feast™ (Ham) 2730 1180 133 56 3.5 315 6600 252 13 15 133 a a a 5 Meat Feast (Canadian Bacon) 2740 1190 134 56 3.5 320 6740 252 13 15 134 a a a Ultimate Supreme 2410 920 104 44 2.5 240 5450 258 15 17 114 a a a 3 Meat Treat® 2760 1230 139 58 3 310 6300 252 13 14 130 a a a Hula Hawaiian® (Ham) 2160 610 69 32 2 200 4960 272 14 30 114 a a a Hula Hawaiian (Canadian Bacon) 2200 640 72 33 2 215 5330 273 13 31 116 a a a Veggie 2250 740 84 39 2 155 5310 266 20 19 101 a a a Large Classic Pizzas Pepperoni 2210 790 90 39 2.5 210 4660 249 13 13 105 a a a Cheese 1950 580 65 31 2 150 3680 248 12 12 95 a a a Sausage 2160 750 85 37 2 190 4350 249 12 12 104 a a a Beef 2150 740 83 38 3 195 4350 248 13 12 106 a a a DEEP!DEEP!™ Dish Pizzas Pepperoni 2770 980 111 47 3 240 5220 319 16 16 129 a a Cheese 2500 750 85 38 2.5 175 4170 317 16 14 118 a a Sausage 2810 1010 114 48 2.5 235 5170 319 16 14 131 a a Beef 2750 950 107 47 3.5 230 5010 318 16 14 130 a a DEEP!DEEP!™ Dish Specialty Pizzas Ultimate Supreme 3050 -

Analysis of Technological and Consumption Quality of Offal And
animals Article Analysis of Technological and Consumption Quality of Offal and Offal Products Obtained from Pulawska and Polish Landrace Pigs Marek Babicz 1 , Kinga Kropiwiec-Doma ´nska 1,* , Ewa Skrzypczak 2 , Magdalena Szyndler-N˛edza 3 and Karolina Szulc 2 1 Institute of Animal Breeding and Biodiversity Conservation, Faculty of Biology, Animal Sciences and Bioeconomy, University of Life Sciences in Lublin, Akademicka 13, 20-950 Lublin, Poland; [email protected] 2 Department of Animal Breeding and Product Quality Assessment, Pozna´nUniversity of Life Sciences, Złotniki, ul Słoneczna 1, 62-002 Suchy Las, Poland; [email protected] (E.S.); [email protected] (K.S.) 3 Department of Pig Breeding, National Research Institute of Animal Production, Krakowska 1, Balice n., 32-083 Kraków, Poland; [email protected] * Correspondence: [email protected] Received: 18 April 2020; Accepted: 29 May 2020; Published: 1 June 2020 Simple Summary: In many countries, offal is an important culinary and technological raw material used for the production of offal dishes and products. Given the growing popularity of such products, as well as insufficient knowledge about the technological and nutritional value of offal, it is important to determine the properties of some pork offal and offal products. The study material consisted of 100 fattening pigs: 50 Pulawska pigs and 50 Polish Landrace pigs. The offal components (tongue, heart, lungs, liver, kidneys) were analysed for physical traits, basic chemical composition and energy value. Offal products (pate, liver sausage, brawn) were made from the offal and their physical, chemical and organoleptic parameters were evaluated. -

Stornoway Black Pudding” EC No: PDO ( ) PGI (D)
SPECIFICATION COUNCIL REGULATION (EC) No 510/2006 on protected geographical indications and protected designations of origin “Stornoway Black Pudding” EC No: PDO ( ) PGI (D) This document sets out the main elements of the product specification for information purposes. 1 RESPONSIBLE DEPARTMENT IN THE MEMBER STATE EU Food Policy Team – Food and Policy Unit Area 7e, 9 Millbank c/o Nobel House 17 Smith Square London SW1P 3JR United Kingdom Tel: +44207 238 6075 Fax: +44207 238 5728 Email: [email protected] 2 GROUP Name: Stornoway Black Pudding Producers’ Association Contact: Claire Macleod, Group Secretary Address: c/o Charles Macleod Limited Ropework Park Matheson Rd Stornoway Isle of Lewis HS1 2LB Tel: 01851 703005 or 07896 897 588 Fax: 01851 704445 E-mail: [email protected] Composition: Producers/processors ( X ) Other ( ) 3 TYPE OF PRODUCT Class 1.2 Meat Products (cooked, salted, smoked) 4 SPECIFICATION (summary of requirements under Article 4(2) of Regulation (EC) No 510/2006) 4.1 Name: “Stornoway Black Pudding” 4.2 Description: Stornoway Black Puddings are a black pudding unique to Stornoway, the capital of the Isle of Lewis in the Outer Hebrides of Scotland. They have a rich, deep reddish-brown to deep brown colour when raw, varying according to individual local recipes. While, according to tradition and heritage, there is some individual variation in the recipes used, the following ingredients are used in the production of Stornoway Black Pudding: • Beef suet • Oatmeal • Onion • Sheep or cow or pig’s blood • Water – where dried blood is used • Salt • Pepper • Skins or casings No other seasonings are permitted and Stornoway Black Puddings must be free from artificial colours, flavours, bulking agents and preservatives. -

Fresh and Fully Cooked Sausage Cooked Salumi
Fresh and Fully Cooked Sausage GUIDELINES FOR STORAGE AND SERVING Storage Sausages should be refrigerated upon receipt until ready to cook. Keep refrigerated below 40° F. Shelf Life Fresh sausages should be used or frozen within 3 days. They may be stored frozen for up to 30 days. Fully cooked sausages are marked with a “use or freeze by” date and may be stored frozen for up to 4 months. Once opened, they should be used within 5 days. A Note on Our Packaging Our sausages are shipped fresh, never frozen, in a Styrofoam cooler with gel ice. Upon arrival, the sausages should be cool to the touch. Typically, the gel ice will be cool and no longer frozen. We use uncoated cardboard boxes and kraft paper, which are fully recyclable. Gel ice and Styrofoam coolers are convenient for re-use. Cooked Salumi GUIDELINES FOR STORAGE AND SERVING Storage Keep refrigerated below 40° F in its original packaging until ready to serve. To preserve freshness, once opened, wrap airtight. Change the wrapping upon each successive opening. Serving Suggestions Slice Classic Mortadella, Salame Rosa, Rosemary Ham, Little Ham or Turkey Galantine paper thin and serve on a platter, or wrap around light breadsticks. Delicious with olives, pickled vegetables, or in a sandwich. A Note on Our Packaging Our cooked salumi products are shipped fresh in a Styrofoam cooler with gel ice. Upon arrival, the product should be cool to the touch. Typically, the gel ice will be cool and no longer frozen. We use uncoated cardboard boxes and kraft paper, which are fully recyclable. -

SAUSAGE Low Risk
SAUSAGE Low Risk ausage comes in many forms, and can contain many Bacteria don’t know whether they are at a 5-star Sdifferent meats—beef, pork, chicken, and turkey. Hot dogs, restaurant, expensive grocery store, or on a local kielbasa, hard pepperoni, hard salami, and bratwurst are all farm—so practice “defensive eating” every time, no types of sausage. For the most part, sausage is low risk, but matter where you get your food, to protect yourself there are still some hazards consumers should be aware of. and your family. Pregnant women should be especially careful of hot dogs, since they can carry Listeria bacteria that can cause miscarriage. Use these tips to avoid illnesses from sausage. Food Preparation Want to know how sausage is made? • Wear disposable plastic gloves when handling raw meat. Sausages and hot dogs use the same leftover meats also Whether or not you wear gloves, wash hands often when found in ground beef. First, meat trimmings are removed handling raw meat. Make sure you clean your hands and from the bones using high pressure or other advanced gloves thoroughly by washing with warm water and soap techniques. Because of concerns about Bovine Spongiform for at least 20 seconds. Encephalopathy (BSE, or “mad cow disease”), for beef • Do not use the same plate, cutting board, other kitchen sausages the process has been refined to ensure that no surface or utensil for raw sausage that is used for cooked bones are included. For poultry, chicken and turkey bones meat or fixings. Wash and sanitize all utensils and surfaces can be pulverized or ground as part of the process, and the that touch raw meat immediately.