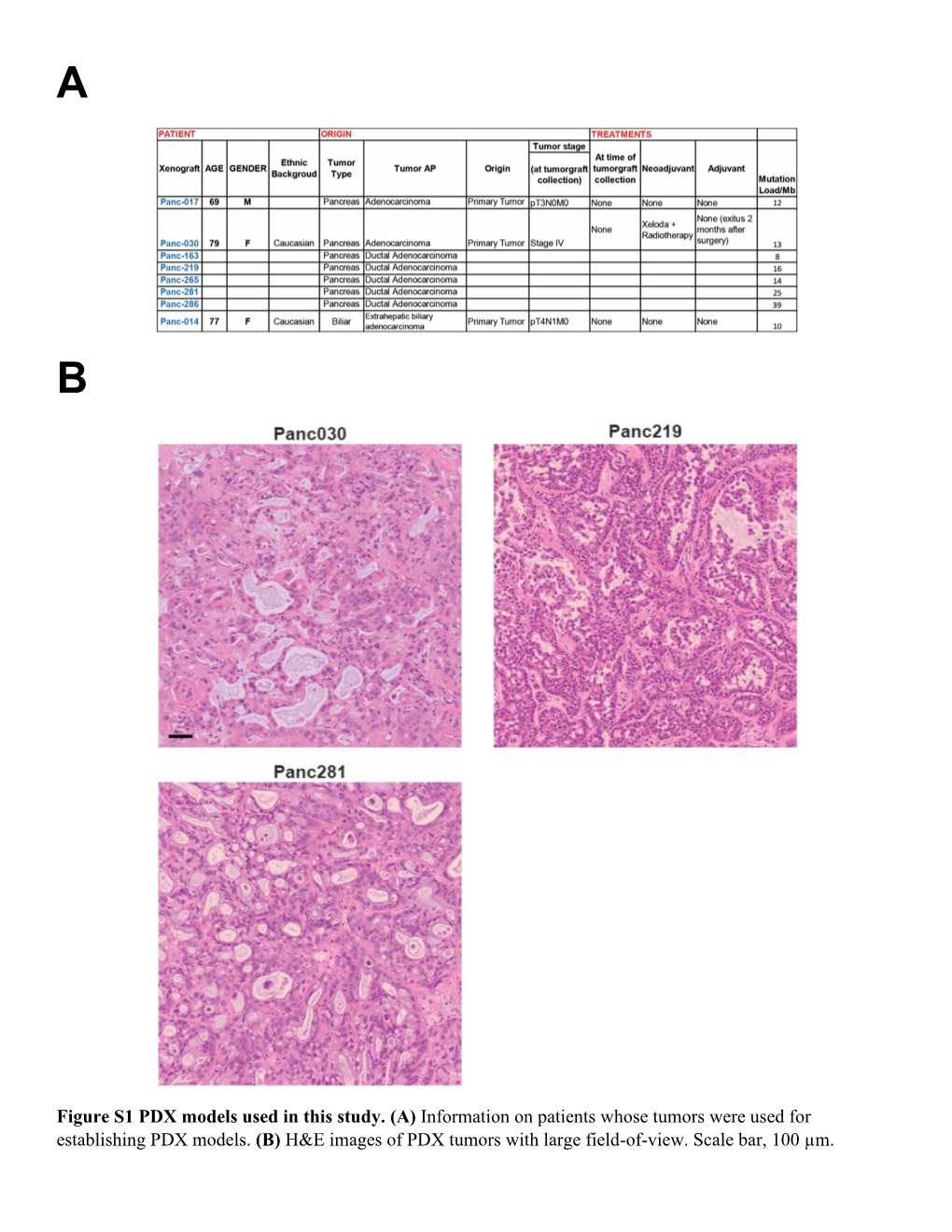
(A) Information on Patients Whose Tumors Were Used for Establishing PDX Models
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Chapter 7: Monogenic Forms of Diabetes
CHAPTER 7 MONOGENIC FORMS OF DIABETES Mark A. Sperling, MD, and Abhimanyu Garg, MD Dr. Mark A. Sperling is Emeritus Professor and Chair, University of Pittsburgh, Department of Pediatrics, Children’s Hospital of Pittsburgh of UPMC, Pittsburgh, PA. Dr. Abhimanyu Garg is Professor of Internal Medicine and Chief of the Division of Nutrition and Metabolic Diseases at University of Texas Southwestern Medical Center, Dallas, TX. SUMMARY Types 1 and 2 diabetes have multiple and complex genetic influences that interact with environmental triggers, such as viral infections or nutritional excesses, to result in their respective phenotypes: young, lean, and insulin-dependence for type 1 diabetes patients or older, overweight, and often manageable by lifestyle interventions and oral medications for type 2 diabetes patients. A small subset of patients, comprising ~2%–3% of all those diagnosed with diabetes, may have characteristics of either type 1 or type 2 diabetes but have single gene defects that interfere with insulin production, secretion, or action, resulting in clinical diabetes. These types of diabetes are known as MODY, originally defined as maturity-onset diabetes of youth, and severe early-onset forms, such as neonatal diabetes mellitus (NDM). Defects in genes involved in adipocyte development, differentiation, and death pathways cause lipodystrophy syndromes, which are also associated with insulin resistance and diabetes. Although these syndromes are considered rare, more awareness of these disorders and increased availability of genetic testing in clinical and research laboratories, as well as growing use of next generation, whole genome, or exome sequencing for clinically challenging phenotypes, are resulting in increased recognition. A correct diagnosis of MODY, NDM, or lipodystrophy syndromes has profound implications for treatment, genetic counseling, and prognosis. -

Downregulation of GNA14 in Hepatocellular Carcinoma Indicates an Unfavorable Prognosis
ONCOLOGY LETTERS 20: 165-172, 2020 Downregulation of GNA14 in hepatocellular carcinoma indicates an unfavorable prognosis TAO YU1*, SIYU LU2* and WENJING XIE2 Departments of 1Medical Oncology and 2Anesthesiology, Xuzhou Municipal Hospital Affiliated to Xuzhou Medical University, Xuzhou, Jiangsu 221000, P.R. China Received August 2, 2019; Accepted March 5, 2020 DOI: 10.3892/ol.2020.11538 Abstract. Guanine nucleotide-binding protein subunit α14 In conclusion, low GNA14 expression may be a novel biomarker (GNA14) knockdown was demonstrated to inhibit the prolifera- for diagnosis and prognosis prediction for patients with HCC. tion of endometrial carcinoma cells in a recent study; however, its role in hepatocellular carcinoma (HCC) is unknown. In Introduction the present study, the clinical significance of GNA14 in HCC was assessed using a dataset of patients with HCC from The Liver cancer was the second leading cause of cancer-asso- Cancer Genome Atlas database. The Integrative Molecular ciated mortality worldwide in 2015 (1). Patients with HCC Database of Hepatocellular Carcinoma and Oncomine data- have no noticeable symptoms, making an accurate diagnosis bases were also used to identify the expression levels of GNA14 challenging; therefore, effective and efficient treatment of in HCC tissues. The association between GNA14 expression HCC should be available at a much earlier stage, and novel levels and clinicopathological features was assessed using the biomarkers are required to improve earlier diagnosis of HCC Wilcoxon signed-rank test and logistic regression analysis. and guide clinical management (2,3). Kaplan-Meier curves and Cox regression analysis were applied HCC is associated with increased expression levels of to evaluate the independent risk factors for clinical outcomes. -

Unnatural Verticilide Enantiomer Inhibits Type 2 Ryanodine Receptor-Mediated Calcium Leak and Is Antiarrhythmic
Unnatural verticilide enantiomer inhibits type 2 ryanodine receptor-mediated calcium leak and is antiarrhythmic Suzanne M. Batistea,1, Daniel J. Blackwellb,1, Kyungsoo Kimb,1, Dmytro O. Kryshtalb, Nieves Gomez-Hurtadob, Robyn T. Rebbeckc, Razvan L. Corneac, Jeffrey N. Johnstona,2, and Bjorn C. Knollmannb,2 aDepartment of Chemistry, Vanderbilt University, Nashville, TN 37235; bDepartment of Medicine, Vanderbilt University Medical Center, Nashville, TN 37232; and cDepartment of Biochemistry, Molecular Biology, and Biophysics, University of Minnesota, Minneapolis, MN 55455 Edited by Dale L. Boger, The Scripps Research Institute, La Jolla, CA, and approved January 15, 2019 (received for review September 27, 2018) Ca2+ leak via ryanodine receptor type 2 (RyR2) can cause poten- heart diseases associated with both atrial and ventricular arrhyth- tially fatal arrhythmias in a variety of heart diseases and has also mia (9). Mutations in RyR2 and its binding partners, which increase + been implicated in neurodegenerative and seizure disorders, mak- SR Ca2 leak, cause primary atrial and ventricular arrhythmia ing RyR2 an attractive therapeutic target for drug development. syndromes such as catecholaminergic polymorphic ventricular Here we synthesized and investigated the fungal natural product tachycardia (CPVT), providing strong evidence for the mechanistic and known insect RyR antagonist (−)-verticilide and several conge- contribution of RyR2 to arrhythmia risk in humans (10). Further ners to determine their activity against mammalian RyR2. Although support comes from gene-targeted mouse models of CPVT, where + the cyclooligomeric depsipeptide natural product (−)-verticilide had catecholamine-induced spontaneous Ca2 release from the SR no effect, its nonnatural enantiomer [ent-(+)-verticilide] signifi- via RyR2 generates potentially fatal cardiac arrhythmias (11, 12). -

Faith and the Human Genome
Plenary Presenters Faith and the Human Genome Faith and the Human Genome Francis S. Collins Despite the best efforts of the American Scientific Affiliation to bridge the gap between science and faith, few gatherings of scientists involved in biology include any meaningful discussion about the spiritual significance of the current revolution in genetics and genomics. Most biologists and geneticists seem to have concluded that science and faith are incompatible, but few who embrace that conclusion seem to have seriously considered the evidence. From my perspective as director of the Human Genome Project, the scientific and religious world views are not only compatible but also inherently complementary. Hence the profound polarization of the scientific and religious perspectives, now glaringly apparent in the fields of biology and genetics, is a source of great distress. Hard-liners in either camp paint increasingly uncompromising pictures that force sincere seekers to choose one view over the other. How all of this must break God’s heart! The elegance and complexity of the human genome is a source of profound wonder. That wonder only strengthens my faith, as it provides glimpses of aspects of From my humanity, which God has known all along, but which we are just now beginning to discover. perspective as e are just on the edge of a whole You made him a little lower than the heav- director of the Whost of developments spurred on enly beings and crowned him with glory by genetics that are going to and honor. You made him ruler over the Human require careful and deliberative thought. -

Epidemiology of Mucopolysaccharidoses Update
diagnostics Review Epidemiology of Mucopolysaccharidoses Update Betul Celik 1,2 , Saori C. Tomatsu 2 , Shunji Tomatsu 1 and Shaukat A. Khan 1,* 1 Nemours/Alfred I. duPont Hospital for Children, Wilmington, DE 19803, USA; [email protected] (B.C.); [email protected] (S.T.) 2 Department of Biological Sciences, University of Delaware, Newark, DE 19716, USA; [email protected] * Correspondence: [email protected]; Tel.: +302-298-7335; Fax: +302-651-6888 Abstract: Mucopolysaccharidoses (MPS) are a group of lysosomal storage disorders caused by a lysosomal enzyme deficiency or malfunction, which leads to the accumulation of glycosaminoglycans in tissues and organs. If not treated at an early stage, patients have various health problems, affecting their quality of life and life-span. Two therapeutic options for MPS are widely used in practice: enzyme replacement therapy and hematopoietic stem cell transplantation. However, early diagnosis of MPS is crucial, as treatment may be too late to reverse or ameliorate the disease progress. It has been noted that the prevalence of MPS and each subtype varies based on geographic regions and/or ethnic background. Each type of MPS is caused by a wide range of the mutational spectrum, mainly missense mutations. Some mutations were derived from the common founder effect. In the previous study, Khan et al. 2018 have reported the epidemiology of MPS from 22 countries and 16 regions. In this study, we aimed to update the prevalence of MPS across the world. We have collected and investigated 189 publications related to the prevalence of MPS via PubMed as of December 2020. In total, data from 33 countries and 23 regions were compiled and analyzed. -

Isoform-Specific Monobody Inhibitors of Small Ubiquitin-Related Modifiers Engineered Using Structure-Guided Library Design
Isoform-specific monobody inhibitors of small ubiquitin-related modifiers engineered using structure-guided library design Ryan N. Gilbretha, Khue Truongb, Ikenna Madub, Akiko Koidea, John B. Wojcika, Nan-Sheng Lia, Joseph A. Piccirillia,c, Yuan Chenb, and Shohei Koidea,1 aDepartment of Biochemistry and Molecular Biology, and cDepartment of Chemistry, University of Chicago, 929 East 57th Street, Chicago, IL 60637; and bDepartment of Molecular Medicine, Beckman Research Institute of the City of Hope, 1450 East Duarte Road, Duarte, CA 91010 Edited by David Baker, University of Washington, Seattle, WA, and approved March 16, 2011 (received for review February 10, 2011) Discriminating closely related molecules remains a major challenge which SUMOylation alters protein function appears to be in the engineering of binding proteins and inhibitors. Here we through SUMO-mediated interactions with other proteins con- report the development of highly selective inhibitors of small ubi- taining a short peptide motif known as a SUMO-interacting motif quitin-related modifier (SUMO) family proteins. SUMOylation is (SIM) (4, 7, 8). involved in the regulation of diverse cellular processes. Functional There are few inhibitors of SUMO/SIM interactions, a defi- differences between two major SUMO isoforms in humans, SUMO1 ciency that limits our ability to finely dissect SUMO biology. In and SUMO2∕3, are thought to arise from distinct interactions the only reported example of such an inhibitor, a SIM-containing mediated by each isoform with other proteins containing SUMO- linear peptide was used to inhibit SUMO/SIM interactions, estab- interacting motifs (SIMs). However, the roles of such isoform- lishing their importance in coordinating DNA repair by nonho- specific interactions are largely uncharacterized due in part to the mologous end joining (9). -

Association of Serum S100B, S100A1 and Zinc-Α2
Progress in Nutrition 2019; Vol. 21, Supplement 1: 154-162 DOI: 10.23751/pn.v21i1-S.5846 © Mattioli 1885 Original articles Association of serum S100B, S100A1 and Zinc-α2- Glycoprotein levels with anthropometric, metabolic and clinical indices in men and women Sorayya Kheirouri1, Mohammad Alizadeh1, Elham Ebrahimi2, Masoumeh Jabbari2 1Associate Professor, Department of Nutrition, Tabriz University of Medical Sciences, Tabriz, Iran - Email: kheirouris@tbzmed. ac.ir, [email protected]; 2MSc. student, Department of Nutrition, Tabriz University of Medical Sciences, Tabriz, Iran Summary. Objectives: We aimed to investigate serum levels of S100B, S100A1, and Zinc-α2- glycoprotein (ZAG) in men and women and to find association of these proteins with anthropometric, metabolic and clini- cal indices. Methods: Eighty-eight apparently healthy adults, 43 men and 45 women, participated in the study. The participants’ body mass index (BMI), waist circumference (WC), systolic and diastolic blood pressure (SBP and DBP) were measured. Serum levels of total cholesterol (TC), triglyceride (TG), low and high den- sity lipoprotein cholesterol (LDL-C and HDL-C), fasting blood sugar (FBS), insulin, S100B, S100A1 and ZAG protein were examined by enzymatic and ELISA laboratory methods. Homeostatic model assessment- insulin resistance (HOMA-IR) index was calculated. Results: Serum levels of S100B, S100A1 and ZAG were comparable between men and women groups. S100B protein was positively associated with TG (r= 0.41, p= 0.006), SBP (r= 0.46, p= 0.002), and DBP (r= 0.37, p= 0.02), but negatively with HDL-c in men. Serum levels of S100A1 were significantly and negatively correlated with WC (r= -0.33, p= 0.03), TG (r= -0.37, p= 0.01), insulin (r= -0.31, p= 0.04) and HOMA-IR (r= -0.32, p= 0.03), in women. -
![Computational Genome-Wide Identification of Heat Shock Protein Genes in the Bovine Genome [Version 1; Peer Review: 2 Approved, 1 Approved with Reservations]](https://docslib.b-cdn.net/cover/8283/computational-genome-wide-identification-of-heat-shock-protein-genes-in-the-bovine-genome-version-1-peer-review-2-approved-1-approved-with-reservations-88283.webp)
Computational Genome-Wide Identification of Heat Shock Protein Genes in the Bovine Genome [Version 1; Peer Review: 2 Approved, 1 Approved with Reservations]
F1000Research 2018, 7:1504 Last updated: 08 AUG 2021 RESEARCH ARTICLE Computational genome-wide identification of heat shock protein genes in the bovine genome [version 1; peer review: 2 approved, 1 approved with reservations] Oyeyemi O. Ajayi1,2, Sunday O. Peters3, Marcos De Donato2,4, Sunday O. Sowande5, Fidalis D.N. Mujibi6, Olanrewaju B. Morenikeji2,7, Bolaji N. Thomas 8, Matthew A. Adeleke 9, Ikhide G. Imumorin2,10,11 1Department of Animal Breeding and Genetics, Federal University of Agriculture, Abeokuta, Nigeria 2International Programs, College of Agriculture and Life Sciences, Cornell University, Ithaca, NY, 14853, USA 3Department of Animal Science, Berry College, Mount Berry, GA, 30149, USA 4Departamento Regional de Bioingenierias, Tecnologico de Monterrey, Escuela de Ingenieria y Ciencias, Queretaro, Mexico 5Department of Animal Production and Health, Federal University of Agriculture, Abeokuta, Nigeria 6Usomi Limited, Nairobi, Kenya 7Department of Animal Production and Health, Federal University of Technology, Akure, Nigeria 8Department of Biomedical Sciences, Rochester Institute of Technology, Rochester, NY, 14623, USA 9School of Life Sciences, University of KwaZulu-Natal, Durban, 4000, South Africa 10School of Biological Sciences, Georgia Institute of Technology, Atlanta, GA, 30032, USA 11African Institute of Bioscience Research and Training, Ibadan, Nigeria v1 First published: 20 Sep 2018, 7:1504 Open Peer Review https://doi.org/10.12688/f1000research.16058.1 Latest published: 20 Sep 2018, 7:1504 https://doi.org/10.12688/f1000research.16058.1 Reviewer Status Invited Reviewers Abstract Background: Heat shock proteins (HSPs) are molecular chaperones 1 2 3 known to bind and sequester client proteins under stress. Methods: To identify and better understand some of these proteins, version 1 we carried out a computational genome-wide survey of the bovine 20 Sep 2018 report report report genome. -

Organ Level Protein Networks As a Reference for the Host Effects of the Microbiome
Downloaded from genome.cshlp.org on October 6, 2021 - Published by Cold Spring Harbor Laboratory Press 1 Organ level protein networks as a reference for the host effects of the microbiome 2 3 Robert H. Millsa,b,c,d, Jacob M. Wozniaka,b, Alison Vrbanacc, Anaamika Campeaua,b, Benoit 4 Chassainge,f,g,h, Andrew Gewirtze, Rob Knightc,d, and David J. Gonzaleza,b,d,# 5 6 a Department of Pharmacology, University of California, San Diego, California, USA 7 b Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California, San Diego, 8 California, USA 9 c Department of Pediatrics, and Department of Computer Science and Engineering, University of 10 California, San Diego California, USA 11 d Center for Microbiome Innovation, University of California, San Diego, California, USA 12 e Center for Inflammation, Immunity and Infection, Institute for Biomedical Sciences, Georgia State 13 University, Atlanta, GA, USA 14 f Neuroscience Institute, Georgia State University, Atlanta, GA, USA 15 g INSERM, U1016, Paris, France. 16 h Université de Paris, Paris, France. 17 18 Key words: Microbiota, Tandem Mass Tags, Organ Proteomics, Gnotobiotic Mice, Germ-free Mice, 19 Protein Networks, Proteomics 20 21 # Address Correspondence to: 22 David J. Gonzalez, PhD 23 Department of Pharmacology and Pharmacy 24 University of California, San Diego 25 La Jolla, CA 92093 26 E-mail: [email protected] 27 Phone: 858-822-1218 28 1 Downloaded from genome.cshlp.org on October 6, 2021 - Published by Cold Spring Harbor Laboratory Press 29 Abstract 30 Connections between the microbiome and health are rapidly emerging in a wide range of 31 diseases. -

Novel Gene Fusions in Glioblastoma Tumor Tissue and Matched Patient Plasma
cancers Article Novel Gene Fusions in Glioblastoma Tumor Tissue and Matched Patient Plasma 1, 1, 1 1 1 Lan Wang y, Anudeep Yekula y, Koushik Muralidharan , Julia L. Small , Zachary S. Rosh , Keiko M. Kang 1,2, Bob S. Carter 1,* and Leonora Balaj 1,* 1 Department of Neurosurgery, Massachusetts General Hospital and Harvard Medical School, Boston, MA 02115, USA; [email protected] (L.W.); [email protected] (A.Y.); [email protected] (K.M.); [email protected] (J.L.S.); [email protected] (Z.S.R.); [email protected] (K.M.K.) 2 School of Medicine, University of California San Diego, San Diego, CA 92092, USA * Correspondence: [email protected] (B.S.C.); [email protected] (L.B.) These authors contributed equally. y Received: 11 March 2020; Accepted: 7 May 2020; Published: 13 May 2020 Abstract: Sequencing studies have provided novel insights into the heterogeneous molecular landscape of glioblastoma (GBM), unveiling a subset of patients with gene fusions. Tissue biopsy is highly invasive, limited by sampling frequency and incompletely representative of intra-tumor heterogeneity. Extracellular vesicle-based liquid biopsy provides a minimally invasive alternative to diagnose and monitor tumor-specific molecular aberrations in patient biofluids. Here, we used targeted RNA sequencing to screen GBM tissue and the matched plasma of patients (n = 9) for RNA fusion transcripts. We identified two novel fusion transcripts in GBM tissue and five novel fusions in the matched plasma of GBM patients. The fusion transcripts FGFR3-TACC3 and VTI1A-TCF7L2 were detected in both tissue and matched plasma. -

Defining Functional Interactions During Biogenesis of Epithelial Junctions
ARTICLE Received 11 Dec 2015 | Accepted 13 Oct 2016 | Published 6 Dec 2016 | Updated 5 Jan 2017 DOI: 10.1038/ncomms13542 OPEN Defining functional interactions during biogenesis of epithelial junctions J.C. Erasmus1,*, S. Bruche1,*,w, L. Pizarro1,2,*, N. Maimari1,3,*, T. Poggioli1,w, C. Tomlinson4,J.Lees5, I. Zalivina1,w, A. Wheeler1,w, A. Alberts6, A. Russo2 & V.M.M. Braga1 In spite of extensive recent progress, a comprehensive understanding of how actin cytoskeleton remodelling supports stable junctions remains to be established. Here we design a platform that integrates actin functions with optimized phenotypic clustering and identify new cytoskeletal proteins, their functional hierarchy and pathways that modulate E-cadherin adhesion. Depletion of EEF1A, an actin bundling protein, increases E-cadherin levels at junctions without a corresponding reinforcement of cell–cell contacts. This unexpected result reflects a more dynamic and mobile junctional actin in EEF1A-depleted cells. A partner for EEF1A in cadherin contact maintenance is the formin DIAPH2, which interacts with EEF1A. In contrast, depletion of either the endocytic regulator TRIP10 or the Rho GTPase activator VAV2 reduces E-cadherin levels at junctions. TRIP10 binds to and requires VAV2 function for its junctional localization. Overall, we present new conceptual insights on junction stabilization, which integrate known and novel pathways with impact for epithelial morphogenesis, homeostasis and diseases. 1 National Heart and Lung Institute, Faculty of Medicine, Imperial College London, London SW7 2AZ, UK. 2 Computing Department, Imperial College London, London SW7 2AZ, UK. 3 Bioengineering Department, Faculty of Engineering, Imperial College London, London SW7 2AZ, UK. 4 Department of Surgery & Cancer, Faculty of Medicine, Imperial College London, London SW7 2AZ, UK. -

Abstracts from the 9Th Biennial Scientific Meeting of The
International Journal of Pediatric Endocrinology 2017, 2017(Suppl 1):15 DOI 10.1186/s13633-017-0054-x MEETING ABSTRACTS Open Access Abstracts from the 9th Biennial Scientific Meeting of the Asia Pacific Paediatric Endocrine Society (APPES) and the 50th Annual Meeting of the Japanese Society for Pediatric Endocrinology (JSPE) Tokyo, Japan. 17-20 November 2016 Published: 28 Dec 2017 PS1 Heritable forms of primary bone fragility in children typically lead to Fat fate and disease - from science to global policy a clinical diagnosis of either osteogenesis imperfecta (OI) or juvenile Peter Gluckman osteoporosis (JO). OI is usually caused by dominant mutations affect- Office of Chief Science Advsor to the Prime Minister ing one of the two genes that code for two collagen type I, but a re- International Journal of Pediatric Endocrinology 2017, 2017(Suppl 1):PS1 cessive form of OI is present in 5-10% of individuals with a clinical diagnosis of OI. Most of the involved genes code for proteins that Attempts to deal with the obesity epidemic based solely on adult be- play a role in the processing of collagen type I protein (BMP1, havioural change have been rather disappointing. Indeed the evidence CREB3L1, CRTAP, LEPRE1, P4HB, PPIB, FKBP10, PLOD2, SERPINF1, that biological, developmental and contextual factors are operating SERPINH1, SEC24D, SPARC, from the earliest stages in development and indeed across generations TMEM38B), or interfere with osteoblast function (SP7, WNT1). Specific is compelling. The marked individual differences in the sensitivity to the phenotypes are caused by mutations in SERPINF1 (recessive OI type obesogenic environment need to be understood at both the individual VI), P4HB (Cole-Carpenter syndrome) and SEC24D (‘Cole-Carpenter and population level.