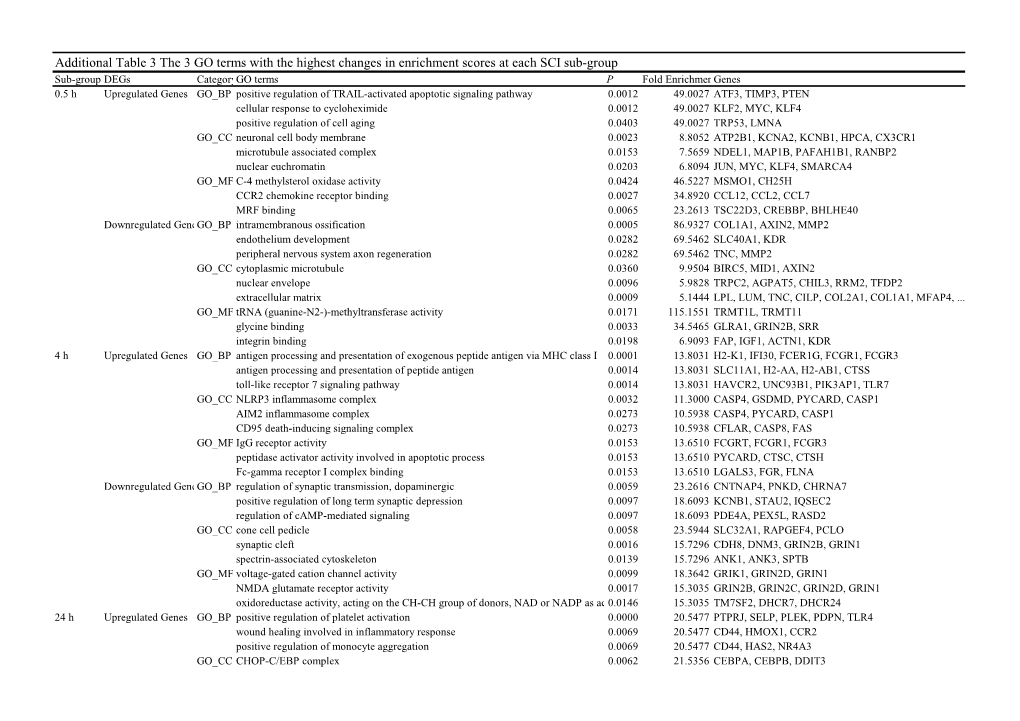
Additional Table 3 the 3 GO Terms with the Highest Changes In
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Mineralocorticoid Receptor Leads to Increased Expression of EGFR
www.nature.com/scientificreports OPEN The mineralocorticoid receptor leads to increased expression of EGFR and T‑type calcium channels that support HL‑1 cell hypertrophy Katharina Stroedecke1,2, Sandra Meinel1,2, Fritz Markwardt1, Udo Kloeckner1, Nicole Straetz1, Katja Quarch1, Barbara Schreier1, Michael Kopf1, Michael Gekle1 & Claudia Grossmann1* The EGF receptor (EGFR) has been extensively studied in tumor biology and recently a role in cardiovascular pathophysiology was suggested. The mineralocorticoid receptor (MR) is an important efector of the renin–angiotensin–aldosterone‑system and elicits pathophysiological efects in the cardiovascular system; however, the underlying molecular mechanisms are unclear. Our aim was to investigate the importance of EGFR for MR‑mediated cardiovascular pathophysiology because MR is known to induce EGFR expression. We identifed a SNP within the EGFR promoter that modulates MR‑induced EGFR expression. In RNA‑sequencing and qPCR experiments in heart tissue of EGFR KO and WT mice, changes in EGFR abundance led to diferential expression of cardiac ion channels, especially of the T‑type calcium channel CACNA1H. Accordingly, CACNA1H expression was increased in WT mice after in vivo MR activation by aldosterone but not in respective EGFR KO mice. Aldosterone‑ and EGF‑responsiveness of CACNA1H expression was confrmed in HL‑1 cells by Western blot and by measuring peak current density of T‑type calcium channels. Aldosterone‑induced CACNA1H protein expression could be abrogated by the EGFR inhibitor AG1478. Furthermore, inhibition of T‑type calcium channels with mibefradil or ML218 reduced diameter, volume and BNP levels in HL‑1 cells. In conclusion the MR regulates EGFR and CACNA1H expression, which has an efect on HL‑1 cell diameter, and the extent of this regulation seems to depend on the SNP‑216 (G/T) genotype. -

Protein Interaction Network of Alternatively Spliced Isoforms from Brain Links Genetic Risk Factors for Autism
ARTICLE Received 24 Aug 2013 | Accepted 14 Mar 2014 | Published 11 Apr 2014 DOI: 10.1038/ncomms4650 OPEN Protein interaction network of alternatively spliced isoforms from brain links genetic risk factors for autism Roser Corominas1,*, Xinping Yang2,3,*, Guan Ning Lin1,*, Shuli Kang1,*, Yun Shen2,3, Lila Ghamsari2,3,w, Martin Broly2,3, Maria Rodriguez2,3, Stanley Tam2,3, Shelly A. Trigg2,3,w, Changyu Fan2,3, Song Yi2,3, Murat Tasan4, Irma Lemmens5, Xingyan Kuang6, Nan Zhao6, Dheeraj Malhotra7, Jacob J. Michaelson7,w, Vladimir Vacic8, Michael A. Calderwood2,3, Frederick P. Roth2,3,4, Jan Tavernier5, Steve Horvath9, Kourosh Salehi-Ashtiani2,3,w, Dmitry Korkin6, Jonathan Sebat7, David E. Hill2,3, Tong Hao2,3, Marc Vidal2,3 & Lilia M. Iakoucheva1 Increased risk for autism spectrum disorders (ASD) is attributed to hundreds of genetic loci. The convergence of ASD variants have been investigated using various approaches, including protein interactions extracted from the published literature. However, these datasets are frequently incomplete, carry biases and are limited to interactions of a single splicing isoform, which may not be expressed in the disease-relevant tissue. Here we introduce a new interactome mapping approach by experimentally identifying interactions between brain-expressed alternatively spliced variants of ASD risk factors. The Autism Spliceform Interaction Network reveals that almost half of the detected interactions and about 30% of the newly identified interacting partners represent contribution from splicing variants, emphasizing the importance of isoform networks. Isoform interactions greatly contribute to establishing direct physical connections between proteins from the de novo autism CNVs. Our findings demonstrate the critical role of spliceform networks for translating genetic knowledge into a better understanding of human diseases. -

Molecular Dissection of G-Protein Coupled Receptor Signaling and Oligomerization
MOLECULAR DISSECTION OF G-PROTEIN COUPLED RECEPTOR SIGNALING AND OLIGOMERIZATION BY MICHAEL RIZZO A Dissertation Submitted to the Graduate Faculty of WAKE FOREST UNIVERSITY GRADUATE SCHOOL OF ARTS AND SCIENCES in Partial Fulfillment of the Requirements for the Degree of DOCTOR OF PHILOSOPHY Biology December, 2019 Winston-Salem, North Carolina Approved By: Erik C. Johnson, Ph.D. Advisor Wayne E. Pratt, Ph.D. Chair Pat C. Lord, Ph.D. Gloria K. Muday, Ph.D. Ke Zhang, Ph.D. ACKNOWLEDGEMENTS I would first like to thank my advisor, Dr. Erik Johnson, for his support, expertise, and leadership during my time in his lab. Without him, the work herein would not be possible. I would also like to thank the members of my committee, Dr. Gloria Muday, Dr. Ke Zhang, Dr. Wayne Pratt, and Dr. Pat Lord, for their guidance and advice that helped improve the quality of the research presented here. I would also like to thank members of the Johnson lab, both past and present, for being valuable colleagues and friends. I would especially like to thank Dr. Jason Braco, Dr. Jon Fisher, Dr. Jake Saunders, and Becky Perry, all of whom spent a great deal of time offering me advice, proofreading grants and manuscripts, and overall supporting me through the ups and downs of the research process. Finally, I would like to thank my family, both for instilling in me a passion for knowledge and education, and for their continued support. In particular, I would like to thank my wife Emerald – I am forever indebted to you for your support throughout this process, and I will never forget the sacrifices you made to help me get to where I am today. -

GABA Receptors
D Reviews • BIOTREND Reviews • BIOTREND Reviews • BIOTREND Reviews • BIOTREND Reviews Review No.7 / 1-2011 GABA receptors Wolfgang Froestl , CNS & Chemistry Expert, AC Immune SA, PSE Building B - EPFL, CH-1015 Lausanne, Phone: +41 21 693 91 43, FAX: +41 21 693 91 20, E-mail: [email protected] GABA Activation of the GABA A receptor leads to an influx of chloride GABA ( -aminobutyric acid; Figure 1) is the most important and ions and to a hyperpolarization of the membrane. 16 subunits with γ most abundant inhibitory neurotransmitter in the mammalian molecular weights between 50 and 65 kD have been identified brain 1,2 , where it was first discovered in 1950 3-5 . It is a small achiral so far, 6 subunits, 3 subunits, 3 subunits, and the , , α β γ δ ε θ molecule with molecular weight of 103 g/mol and high water solu - and subunits 8,9 . π bility. At 25°C one gram of water can dissolve 1.3 grams of GABA. 2 Such a hydrophilic molecule (log P = -2.13, PSA = 63.3 Å ) cannot In the meantime all GABA A receptor binding sites have been eluci - cross the blood brain barrier. It is produced in the brain by decarb- dated in great detail. The GABA site is located at the interface oxylation of L-glutamic acid by the enzyme glutamic acid decarb- between and subunits. Benzodiazepines interact with subunit α β oxylase (GAD, EC 4.1.1.15). It is a neutral amino acid with pK = combinations ( ) ( ) , which is the most abundant combi - 1 α1 2 β2 2 γ2 4.23 and pK = 10.43. -

Transcriptomic Analysis of Native Versus Cultured Human and Mouse Dorsal Root Ganglia Focused on Pharmacological Targets Short
bioRxiv preprint doi: https://doi.org/10.1101/766865; this version posted September 12, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Transcriptomic analysis of native versus cultured human and mouse dorsal root ganglia focused on pharmacological targets Short title: Comparative transcriptomics of acutely dissected versus cultured DRGs Andi Wangzhou1, Lisa A. McIlvried2, Candler Paige1, Paulino Barragan-Iglesias1, Carolyn A. Guzman1, Gregory Dussor1, Pradipta R. Ray1,#, Robert W. Gereau IV2, # and Theodore J. Price1, # 1The University of Texas at Dallas, School of Behavioral and Brain Sciences and Center for Advanced Pain Studies, 800 W Campbell Rd. Richardson, TX, 75080, USA 2Washington University Pain Center and Department of Anesthesiology, Washington University School of Medicine # corresponding authors [email protected], [email protected] and [email protected] Funding: NIH grants T32DA007261 (LM); NS065926 and NS102161 (TJP); NS106953 and NS042595 (RWG). The authors declare no conflicts of interest Author Contributions Conceived of the Project: PRR, RWG IV and TJP Performed Experiments: AW, LAM, CP, PB-I Supervised Experiments: GD, RWG IV, TJP Analyzed Data: AW, LAM, CP, CAG, PRR Supervised Bioinformatics Analysis: PRR Drew Figures: AW, PRR Wrote and Edited Manuscript: AW, LAM, CP, GD, PRR, RWG IV, TJP All authors approved the final version of the manuscript. 1 bioRxiv preprint doi: https://doi.org/10.1101/766865; this version posted September 12, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

A 3-Fold Kernel Approach for Characterizing Late Onset Alzheimer’S Disease
bioRxiv preprint doi: https://doi.org/10.1101/397760; this version posted August 22, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. A 3-fold kernel approach for characterizing Late Onset Alzheimer’s Disease Margherita Squillarioa,*, Federico Tomasia, Veronica Tozzoa, Annalisa Barlaa and Daniela Ubertib “for the Alzheimer’s Disease Neuroimaging Initiative**” aDIBRIS, University of Genoa, Via Dodecaneso 35, I-16146 Genova, Italy. E-mail address: {squillario, federico.tomasi, veronica.tozzo}@dibris.unige.it, [email protected] bDepartment of Molecular and Translational Medicine, University of Brescia, Viale Europa 11, 25123, Brescia, Italy. E-mail address: [email protected] * Corresponding author and Lead Contact. Tel: +39-010-353-6707; Fax: +39-010-353- 6699. ** Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp- content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf 1 bioRxiv preprint doi: https://doi.org/10.1101/397760; this version posted August 22, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

GABA Receptor Crosstalk the Inhibitory Neurotransmitter GABA Activates Both Ionotropic
RESEARCH HIGHLIGHTS IN BRIEF SYNAPTIC TRANSMISSION GABA receptor crosstalk The inhibitory neurotransmitter GABA activates both ionotropic type A GABA receptors (GABAARs) and metabotropic GABAB receptors (GABABRs). Two independent studies in rat brain slices now show that activation of postsynaptic GABABRs enhances signalling through extrasynaptic GABAARs. Activity-dependent changes in GABA concentration activate postsynaptic GABABRs leading to slow inhibition, whereas high-affinity GABAARs at extrasynaptic sites are activated tonically by ambient GABA. This tonic inhibitory tone has been implicated in synaptic integration, anxiety-related behaviours and seizure susceptibility. Tao et al. show that activation of postsynaptic GABABRs enhances GABAAR-mediated currents caused by exogenous GABA or the GABABR agonist baclofen in dentate gyrus granule cells (DGGCs) but not in CA1 pyramidal neurons or cortical layer 2/3 pyramidal neurons. Connelly et al. report similar crosstalk in thalamocortical neurons of the ventrobasal thalamus and cerebellar granule cells as well as in DGGCs. ORIGINAL RESEARCH PAPERS Tao, W. et al. Postsynaptic GABAB receptors enhance extrasynaptic GABAA receptor function in dentate gyrus granule cells. J. Neurosci. 33, 3738–3743 (2013) | Connelly, W. M. et al. GABAB receptors regulate extrasynaptic GABAA receptors. J. Neurosci. 33, 3780–3785 (2013) SIGNAL TRANSDUCTION JACOB reveals the origin of NMDAR signals Signalling through NMDA receptors (NMDARs) can trigger cell death or survival depending on whether activation of such receptors occurs at extrasynaptic or synaptic sites, respectively. It is unclear how these opposing signals are communicated to and discriminated by the nucleus, but the authors of this study show that differential phosphorylation of JACOB relays the origin of the NMDAR signal to the nucleus. -

Epac2a-Null Mice Exhibit Obesity-Prone Nature More Susceptible to Leptin Resistance
OPEN International Journal of Obesity (2017) 41, 279–288 www.nature.com/ijo ORIGINAL ARTICLE Epac2a-null mice exhibit obesity-prone nature more susceptible to leptin resistance M Hwang1,5,YGo2,5,6, J-H Park1, S-K Shin1, SE Song1, B-C Oh3, S-S Im1, I Hwang1, YH Jeon2, I-K Lee2, S Seino4 and D-K Song1 BACKGROUND: The exchange protein directly activated by cAMP (Epac), which is primarily involved in cAMP signaling, has been known to be essential for controlling body energy metabolism. Epac has two isoforms: Epac1 and Epac2. The function of Epac1 on obesity was unveiled using Epac1 knockout (KO) mice. However, the role of Epac2 in obesity remains unclear. METHODS: To evaluate the role of Epac2 in obesity, we used Epac2a KO mice, which is dominantly expressed in neurons and endocrine tissues. Physiological factors related to obesity were analyzed: body weight, fat mass, food intake, plasma leptin and adiponectin levels, energy expenditure, glucose tolerance, and insulin and leptin resistance. To determine the mechanism of Epac2a, mice received exogenous leptin and then hypothalamic leptin signaling was analyzed. RESULTS: Epac2a KO mice appeared to have normal glucose tolerance and insulin sensitivity until 12 weeks of age, but an early onset increase of plasma leptin levels and decrease of plasma adiponectin levels compared with wild-type mice. Acute leptin injection revealed impaired hypothalamic leptin signaling in KO mice. Consistently, KO mice fed a high-fat diet (HFD) were significantly obese, presenting greater food intake and lower energy expenditure. HFD-fed KO mice were also characterized by greater impairment of hypothalamic leptin signaling and by weaker leptin-induced decrease in food consumption compared with HFD-fed wild-type mice. -

Ion Channels 3 1
r r r Cell Signalling Biology Michael J. Berridge Module 3 Ion Channels 3 1 Module 3 Ion Channels Synopsis Ion channels have two main signalling functions: either they can generate second messengers or they can function as effectors by responding to such messengers. Their role in signal generation is mainly centred on the Ca2 + signalling pathway, which has a large number of Ca2+ entry channels and internal Ca2+ release channels, both of which contribute to the generation of Ca2 + signals. Ion channels are also important effectors in that they mediate the action of different intracellular signalling pathways. There are a large number of K+ channels and many of these function in different + aspects of cell signalling. The voltage-dependent K (KV) channels regulate membrane potential and + excitability. The inward rectifier K (Kir) channel family has a number of important groups of channels + + such as the G protein-gated inward rectifier K (GIRK) channels and the ATP-sensitive K (KATP) + + channels. The two-pore domain K (K2P) channels are responsible for the large background K current. Some of the actions of Ca2 + are carried out by Ca2+-sensitive K+ channels and Ca2+-sensitive Cl − channels. The latter are members of a large group of chloride channels and transporters with multiple functions. There is a large family of ATP-binding cassette (ABC) transporters some of which have a signalling role in that they extrude signalling components from the cell. One of the ABC transporters is the cystic − − fibrosis transmembrane conductance regulator (CFTR) that conducts anions (Cl and HCO3 )and contributes to the osmotic gradient for the parallel flow of water in various transporting epithelia. -

A Review of Glutamate Receptors I: Current Understanding of Their Biology
J Toxicol Pathol 2008; 21: 25–51 Review A Review of Glutamate Receptors I: Current Understanding of Their Biology Colin G. Rousseaux1 1Department of Pathology and Laboratory Medicine, Faculty of Medicine, University of Ottawa, Ottawa, Ontario, Canada Abstract: Seventy years ago it was discovered that glutamate is abundant in the brain and that it plays a central role in brain metabolism. However, it took the scientific community a long time to realize that glutamate also acts as a neurotransmitter. Glutamate is an amino acid and brain tissue contains as much as 5 – 15 mM glutamate per kg depending on the region, which is more than of any other amino acid. The main motivation for the ongoing research on glutamate is due to the role of glutamate in the signal transduction in the nervous systems of apparently all complex living organisms, including man. Glutamate is considered to be the major mediator of excitatory signals in the mammalian central nervous system and is involved in most aspects of normal brain function including cognition, memory and learning. In this review, the basic biology of the excitatory amino acids glutamate, glutamate receptors, GABA, and glycine will first be explored. In the second part of this review, the known pathophysiology and pathology will be described. (J Toxicol Pathol 2008; 21: 25–51) Key words: glutamate, glycine, GABA, glutamate receptors, ionotropic, metabotropic, NMDA, AMPA, review Introduction and Overview glycine), peptides (vasopressin, somatostatin, neurotensin, etc.), and monoamines (norepinephrine, dopamine and In the first decades of the 20th century, research into the serotonin) plus acetylcholine. chemical mediation of the “autonomous” (autonomic) Glutamatergic synaptic transmission in the mammalian nervous system (ANS) was an area that received much central nervous system (CNS) was slowly established over a research activity. -

Ion Channels
UC Davis UC Davis Previously Published Works Title THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Ion channels. Permalink https://escholarship.org/uc/item/1442g5hg Journal British journal of pharmacology, 176 Suppl 1(S1) ISSN 0007-1188 Authors Alexander, Stephen PH Mathie, Alistair Peters, John A et al. Publication Date 2019-12-01 DOI 10.1111/bph.14749 License https://creativecommons.org/licenses/by/4.0/ 4.0 Peer reviewed eScholarship.org Powered by the California Digital Library University of California S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2019/20: Ion channels. British Journal of Pharmacology (2019) 176, S142–S228 THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Ion channels Stephen PH Alexander1 , Alistair Mathie2 ,JohnAPeters3 , Emma L Veale2 , Jörg Striessnig4 , Eamonn Kelly5, Jane F Armstrong6 , Elena Faccenda6 ,SimonDHarding6 ,AdamJPawson6 , Joanna L Sharman6 , Christopher Southan6 , Jamie A Davies6 and CGTP Collaborators 1School of Life Sciences, University of Nottingham Medical School, Nottingham, NG7 2UH, UK 2Medway School of Pharmacy, The Universities of Greenwich and Kent at Medway, Anson Building, Central Avenue, Chatham Maritime, Chatham, Kent, ME4 4TB, UK 3Neuroscience Division, Medical Education Institute, Ninewells Hospital and Medical School, University of Dundee, Dundee, DD1 9SY, UK 4Pharmacology and Toxicology, Institute of Pharmacy, University of Innsbruck, A-6020 Innsbruck, Austria 5School of Physiology, Pharmacology and Neuroscience, University of Bristol, Bristol, BS8 1TD, UK 6Centre for Discovery Brain Science, University of Edinburgh, Edinburgh, EH8 9XD, UK Abstract The Concise Guide to PHARMACOLOGY 2019/20 is the fourth in this series of biennial publications. The Concise Guide provides concise overviews of the key properties of nearly 1800 human drug targets with an emphasis on selective pharmacology (where available), plus links to the open access knowledgebase source of drug targets and their ligands (www.guidetopharmacology.org), which provides more detailed views of target and ligand properties. -

Multi-Functionality of Proteins Involved in GPCR and G Protein Signaling: Making Sense of Structure–Function Continuum with In
Cellular and Molecular Life Sciences (2019) 76:4461–4492 https://doi.org/10.1007/s00018-019-03276-1 Cellular andMolecular Life Sciences REVIEW Multi‑functionality of proteins involved in GPCR and G protein signaling: making sense of structure–function continuum with intrinsic disorder‑based proteoforms Alexander V. Fonin1 · April L. Darling2 · Irina M. Kuznetsova1 · Konstantin K. Turoverov1,3 · Vladimir N. Uversky2,4 Received: 5 August 2019 / Revised: 5 August 2019 / Accepted: 12 August 2019 / Published online: 19 August 2019 © Springer Nature Switzerland AG 2019 Abstract GPCR–G protein signaling system recognizes a multitude of extracellular ligands and triggers a variety of intracellular signal- ing cascades in response. In humans, this system includes more than 800 various GPCRs and a large set of heterotrimeric G proteins. Complexity of this system goes far beyond a multitude of pair-wise ligand–GPCR and GPCR–G protein interactions. In fact, one GPCR can recognize more than one extracellular signal and interact with more than one G protein. Furthermore, one ligand can activate more than one GPCR, and multiple GPCRs can couple to the same G protein. This defnes an intricate multifunctionality of this important signaling system. Here, we show that the multifunctionality of GPCR–G protein system represents an illustrative example of the protein structure–function continuum, where structures of the involved proteins represent a complex mosaic of diferently folded regions (foldons, non-foldons, unfoldons, semi-foldons, and inducible foldons). The functionality of resulting highly dynamic conformational ensembles is fne-tuned by various post-translational modifcations and alternative splicing, and such ensembles can undergo dramatic changes at interaction with their specifc partners.