Isolation of Nitrate and Phosphate Removing Bacteria from Various Environmental Sites
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Dreamland Enclave
https://www.propertywala.com/dreamland-enclave-kolkata Dreamland Enclave - Kaikhali, Kolkata Residential Apartments Somani Dreamland Enclave, is Somani's Realtors recent promotion, which can fulfill all your dream of a sweet home. Home, sweet home. Home from where we begin our day. Home, where we like to see sweet dream . Project ID : J602119049 Builder: Somani Realtors Pvt. Ltd. Properties: Apartments / Flats Location: Dreamland Enclave, Kaikhali, Kolkata - 700052 (West Bengal) Completion Date: Feb, 2011 Status: Completed Description Somani Realtors Pvt. Ltd. is one of the leading real estate project marketers in Kolkata. It is a team of marketing experts with the motto of professional service to both-the developers and the customers.This team is in service for one and a half decade and has a reputation par excellence. The company has successfully marketed various projects which are now landmark destinations.For the home seekers, Somani Realtors is known to be one stop hassle-free shop. Somani Dreamland Enclave, is Somani's Realtors recent promotion, which can fulfill all your dream of a sweet home. Home, sweet home. Home from where we begin our day. Home, where we like to see sweet dream . A home is an ideal home where we can live with joy and happiness.In this g+5 multistoried residential apartment every flat is either pool facing or south or garden facing. So from your private balcony you can enjoy the beauty of the nature and can take a pollution free air with full of oxygen.It's an exquisite, unique and filled with all modern day facilities within the periphery of a quality construction, spilt into four blocks, each one is exposed to ample light and fresh air. -

Come, Experience the Bloom
Come,Come, experienceexperience thethe bloom.bloom. About Us Over twenty five years ago a few enterprising men kindled the fire of progress and made excellence their commitment and growth their goal. Accomplishing this noble objective called for indefatigable determination, fine tuned policies, dedicated manpower and world class technology to shape the future. Real Estate Development The principal activity of the GM Group is development of residential and commercial buildings. The Group is well known and recognized by the style and name of GM Group and has been at the helm of the real estate industry for over two decades. Projects span the major Metros and outskirts. The combined real estate value of the projects developed would run into several million Rupees, a substantial sum by any national or international standard. Some of the major landmarks stand proud testimony to the development activities of the Group - Meena Residency, Meena Ganga, Meena Florence, Meena Green are just a few of them. Prestigious completed residential projects in Kolkata are Madhu Apartments, Meena View, Meena Vatika and many more. Ethically, GM Group associates only reputed vendors who have proven track record. As a result buyers need not fear any compromise on the quality of construction when they buy property from us. GM Group having an exceptional database and professional contacts assure a choice of properties to fit in the budget, in Kolkata and the suburbs. E S Nearby N Locations W Airport Delhi Public Eco Park School, Newtown Delhi Public School, Newtown -

Indeed Resume
D BHATTACHARJEE Tutorial comprises an Expert Group of Home Tutors: D Bhattacharjee teaches UNIVERSITY ENGLISH HONOURS MA ENGLISH ICSE ISC CBSE IGCSE ENGLISH TUITION COACHING CLASS TUTORIAL at KESTOPUR KOLKATA Kolkata, West Bengal [email protected] 9836618842 27 YEARS’ EXP. EX SCHOOL TEACHER SUSMITA MADAM 9836618836 TEACHES BENGALI ICSE ISC CBSE WB BOARD COACHING CLASSES TUTORIAL at KESTOPUR, NEW TOWN, SALT LAKE, VIP KOLKATA 35 YRS EXP ENGLISH TUTOR FOR ISC CBSE ICSE MA, BA ENGLISH HONOURS, IGCSE CAMBRIDGE O & A LEVEL, BBA, BCA (ENGLISH ONLY) D BHATTACHARJEE TEACHES AND PROVIDES STUDY MATERIALS HOME TUITION COACHING CLASSES AT KESTOPUR TUTORIAL D BHATTACHARJEE www.dbhattacharjee.com TEACHES ENGLISH HONOURS, MA (ENGLISH) BBA BCA ENGLISH FOR THE STUDENTS FROM THE CALCUTTA UNIVERSITY, IGNOU, NETAJI OPEN UNIVERSITY, VIDYASAGAR UNIVERSITY, RABINDRA BHARATI UNIVERSITY ETC. 9836618842 An SST HISTORY GEOGRAPHY CIVICS tutor teaches visiting students' places at KESTOPUR, NEW TOWN, SALT LAKE areas for ICSE CBSE V-X. WHATSAPP / PH. No. 70441 06207 Willing to relocate to: NEW TOWN, SALT LAKE, VIP - Kolkata, West Bengal - Kolkata, West Bengal Work Experience 35 years' TEACHING ENGLISH LANGUAGE AND LITERATURE English BENGALI science math social science TUITION www.dbhattacharjee.com - Kolkata, West Bengal March 1985 to Present ENGLISH HOME TUTOR D BHATTACHARJEE TEACHES ENGLISH HONOURS, CAMBRIDGE IGCSE, ISC, ICSE, CBSE VII-XII AT STUDENTS' / OWN PLACE. ENGLISH HOME TUITION SLOT (Individually / Group at students’ place ) : ONE HOUR AND A HALF (ONCE A WEEK). ENGLISH TUTORIAL SLOT (In his tutorial Batch ) : THREE HOURS (ONCE A WEEK). FOR MOCK TEST, DOUBT CLEARING & EXAM PREPARATION STUDENTS’ GUARDIANS MAY SEPARATELY CONTACT FOR ENGLISH LANGUAGE LITERATURE NOTES, SOLVED QUESTION - ANSWERS, GUIDELINES, SUGGESTIONS & SAMPLE PAPERS, MODEL TEST PAPERS ETC. -
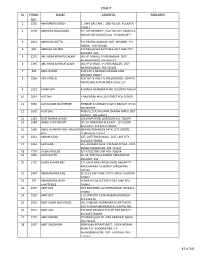
Sl Form No. Name Address Remarks
STARLIT SL FORM NAME ADDRESS REMARKS NO. 1 2235 ABHIMANYU SINGH 2, UMA DAS LANE , 2ND FLOOR , KOLKATA- 700013 2 1998 ABHISHEK MAJUMDER R.P. APPARTMENT , FLAT NO-303 PRAFULLA KANAN (W) KOLKATA-101 P.S-BAGUIATI 3 1922 ABHRANIL DUTTA P.O-PRAFULLANAGAR DIST-24PGS(N) , P.S- HABRA , PIN-743268 4 860 ABINASH HALDER 139 BELGACHIA EAST HB-6 SALT LAKE CITY KOLKATA -106 5 2271 ABU HENA MONIRUL ALAM VILL+P.O-MILKI, P.S-REJINAGAR DIST- MURSHIDABAD, PIN-742163 6 1395 ABU HENA SAHINUR ALAM VILL+P.O-MILKI , P.S-REGINAGAR , DIST- MURSHIDABAD, PIN-742163 7 446 ABUL HASAN B 32 1AH 3 MIAJAN OSTAGAR LANE KOLKATA 700017 8 3286 ADA AFREEN FLAT NO 9I PINES IV GREENWOOD SONATA NEWTOWN ACTION AREA II KOL 157 9 1623 ADHIR GIRI 8 DURGA CHANDRA ROAD KOLKATA 700014 10 2807 AJIT DAS 6 RAJENDRA MULLICK STREET KOL-700007 11 3650 AJIT KUMAR MUKHERJEE SHIBBARI K,S ROAD P.O &P.S NAIHATI DT 24 PGS NORTH 12 1602 AJOY DAS PO&VILL SOUTH GARIA CHARAK MATH DIST 24 PGS S PIN 743613 13 1261 AJOY KUMAR GHOSH 326 JAWPUR RD. DUMDUM KOL-700074 14 1584 AKASH CHOUDHURY DD-19, NARAYANTALA EAST , 1ST FLOOR BAGUIATI , KOLKATA-700059 15 2460 AKHIL CHANDRA ROY MNJUSRI 68 RANI RASHMONI PATH CITY CENTRE ROY DURGAPUR 713216 16 2311 AKRAM AZAD 227, DUTTABAD ROAD, SALT LAKE CITY , KOLKATA-700064 17 1441 ALIP JANA VILL-KHAMAR CHAK P.O-NILKUNTHIA , DIST- PURBA MIDNAPORE PIN-721627 18 2797 ALISHA BEGUM 5/2 B DOCTOR LANE KOL-700014 19 1956 ALOK DUTTA AC-64, PRAFULLA KANAN KRISHNAPUR KOLKATA -101 20 1719 ALOKE KUMAR DEY 271 SASHI BABU ROAD SAHID NAGAR PO KANCHAPARA PS BIZPUR 24PGSN PIN 743145 21 3447 -

BUS ROUTE-18-19 Updated Time.Xls LIST of DROP ROUTES & STOPPAGES TIMINGS LIST of DROP ROUTES & STOPPAGES TIMINGS
LIST OF DROP ROUTES & STOPPAGES TIMINGS LIST OF DROP ROUTES & STOPPAGES TIMINGS FOR THE SESSION 2018-19 FOR THE SESSION 2018-19 ESTIMATED TIMING MAY CHANGE SUBJECT ESTIMATED TIMING MAY CHANGE SUBJECT TO TO CONDITION OF THE ROAD CONDITION OF THE ROAD ROUTE NO - 1 ROUTE NO - 2 SL NO. LIST OF DROP STOPPAGES TIMINGS SL NO. LIST OF DROP STOPPAGES TIMINGS 1 DUMDUM CENTRAL JAIL 13.00 1 IDEAL RESIDENCY 13.05 2 CLIVE HOUSE, MALL ROAD 13.03 2 KANKURGACHI MORE 13.07 3 KAJI PARA 13.05 3 MANICKTALA RAIL BRIDGE 13.08 4 MOTI JEEL 13.07 4 BAGMARI BAZAR 13.10 5 PRIVATE ROAD 13.09 5 MANICKTALA P.S. 13.12 6 CHATAKAL DUMDUM ROAD 13.11 6 MANICKTALA DINENDRA STREET XING 13.14 7 HANUMAN MANDIR 13.13 7 MANICKTALA BLOOD BANK 13.15 8 DUMDUM PHARI 01:15 8 GIRISH PARK METRO STATION 13.20 9 DUMDUM STATION 01:17 9 SOVABAZAR METRO STATION 13.23 10 7 TANK, DUMDUM RD 01:20 10 B.K.PAUL AVENUE 13.25 11 AHIRITALA SITALA MANDIR 13.27 12 JORABAGAN PARK 13.28 13 MALAPARA 13.30 14 GANESH TALKIES 13.32 15 RAM MANDIR 13.34 16 MAHAJATI SADAN 13.37 17 CENTRAL AVENUE RABINDRA BHARATI 13.38 18 M.G.ROAD - C.R.AVENUE XING 13.40 19 MOHD.ALI PARK 13.42 20 MEDICAL COLLEGE 13.44 21 BOWBAZAR XING 13.46 22 INDIAN AIRLINES 13.48 23 HIND CINEMA XING 13.50 24 LEE MEMORIAL SCHOOL - LENIN SR. 13.51 ROUTE NO - 3 ROUTE NO - 04 SL NO. -

List of Eligible Hospitals / Diagnostic Centres for Empanelment Under Cghs Kolkata, 2014 I
LIST OF ELIGIBLE HOSPITALS / DIAGNOSTIC CENTRES FOR EMPANELMENT UNDER CGHS KOLKATA, 2014 I. Hospitals: Kolkata NABH PBG Sl Name of the Hospital Address & Telephone No Accredited/Non Facilities Empanelled for submitted No NABH (Y/N) General Medicine, Non‐ invasive Cardiology, Neurology and Neuro Surgery, Spine and Trauma Surgery, Urology, (excluding renal Transplant), Dermatology, Diabetology, Endocrinolgoy, Orthopedics including joint replacement 276, Canal Street, Kolkata ‐ Daffodil Hospitals Pvt. surgeries , Medical Oncology, 1 700048 Tel. No.‐033‐ NON NABH Yes Ltd (Hospital). ENT, Gastroenterology and 25346649/5681 Gastro Surgery, Neonatology, Paediatrics and Paediatric Surgery, Gynaecology & Obstetrics, Nephrology including Hemo‐dialysis both sero‐positive & sero‐negative, General and Laproscopic Surgeries, Plastic Surgery, Dental 1/1, National Library B.M. Birla Heart Research Cardiology and Cardio‐ 2 Avenue, Kolkata‐700027 Tel. NABH Yes Centre thoracic Surgery No.‐033‐24567890 Cardiology and Cardio‐ thoracic Surgery, Orthopedic with joint replacement, Nephrology , Urology with renal transpantation, Endocrinology, Neuro Premises Number‐ Surgery, Dialysis, Rabindranath Tagore 1489, 124, Dermatology, ENT, 3 International Institute of Mukundapur, EM Bypass, NABH Gastroentrology and GI Yes Cardiac Sciences Kolkata‐700099 Tel. No.‐ Surgery, General Surgery, 033‐7122‐2222 General Medicine, Gynecology and Obstetrics, Haemato‐oncology, Nuclear Medicine, Plastic Surgey, Paediatrics and Neonatology, Paediatric Surgery, Respiratory Medicine -
Pick up Fee with Chart.Xlsx
Pick‐Up Routes with time (NURSERY ‐ CLASS‐X) for The Academic Session 2018‐19 Route # 1 (8585059795) Time KM Fee(INR) Route # 9 (8585014347) Time KM Fee(INR) Route # 15 (8585059823) Time KM Fee(INR) Route # 23 (7603089214) Time KM Fee(INR) Route # 31 (7595064661) Time KM Fee(INR) Lake Town VIP 07:00 AM Above 15 Km 3900 Sealdah ESIC Hospital 06.40 Am Above 15 Km 3900 Newtown Metro Plaza 07.15 Am 0 ‐ 10 Km 3400 Kestopur Subway 07:05 AM 11 Km ‐ 15 Km 3650 Hind Cinema 06:55 AM Above 15 Km 3900 Lake Town Host 07:02 AM Above 15 Km 3900 Rajabazar Sci. College Bus Stop 06.45 Am Above 15 Km 3900 Rajarhat Check Post 07.20 Am 0 ‐ 10 Km 3400 Baguiati College 07:08 AM 11 Km ‐ 15 Km 3650 Chandni Chowk Metro 06:58 AM Above 15 Km 3900 Lake Town Nursing Home 07:04 AM Above 15 Km 3900 Karbala 06.47 Am Above 15 Km 3900 Route # 16 (8585059796) Time KM Fee(INR) Baguiati Bus Stop 07:12 AM 11 Km ‐ 15 Km 3650 C.R Avenue Metro 07:00 AM Above 15 Km 3900 Lake Town Mansa Bari 07:05 AM Above 15 Km 3900 Sahitya Parishad Crossing 06.50 Am Above 15 Km 3900 Park Circus 7 Point 06:40 AM Above 15 Km 3900 Baguiati Big Bazar(opp) 07:15 AM 11 Km ‐ 15 Km 3650 Md. Ali Park 07:05 AM Above 15 Km 3900 Route # 2 (8585059790) Time KM Fee(INR) Pareshnath Mandir Crossing 06.53 Am Above 15 Km 3900 Quest Mall 06:42 AM Above 15 Km 3900 Raghunathpur Bus Stop 07:18 AM 11 Km ‐ 15 Km 3650 Ram Mandir 07:10 AM 11 Km ‐ 15 Km 3650 Bangur Swimming Pool 07:00 AM Above 15 Km 3900 Deshbandhu Park 06.58 Am Above 15 Km 3900 Ballygunge Phari 06:45 AM Above 15 Km 3900 Route # 24 (7603089215) Time KM -

Affected Zone Wise Division of Containment Zones in North 24 Parganas
Affected Zone wise Division of Containment Zones in North 24 Parganas SL No Muni/Block Ward/GP Affected Zone Police Station 1 BSF Camp, Alambazar Baranagar 4 2 Kumarpara Lane,Jhilbagan, Kol-35 Baranagar 3 8 Deshbandhu Road(West) Baranagar 4 13 Gopal Lal Thakur Road, Baranagar Baranagar 5 14 Banhoogly , Rabindranagar Baranagar 6 17 Nabin Chandra Das Road, Sreepalli Baranagar 7 18 A K Mukherjee Road Baranagar A. K Mukherjee Rd 8Baranagar 19 Baranagar Baranagar, Subhas Nagar, Noa para 9 21 Bir Anantaram Mondal Lane Baranagar 10 25 Jogendra Basak Road, Baranagar Baranagar 11 26 Jogendra Basak Road, Baranagar Baranagar 12 27 Ramchand Mukherjee Lane Baranagar 13 32 MNK Road (S) Baranagar 14 33 JN Banerjee lane Baranagar 15 34 B.K. Maitra Road Baranagar 15 Kazipara Barasat 16 Barasat 17 31 Hridaypur Barasat 18 5 Sadhu Mukherjee Road. Titagarh Sumagalpur Old Calcutta Road, Barrackpore, 19 10 Titagarh Barrackpore Talpukur 20 19 Gandhi more near Mother Teresa Hospital Titagarh 21 23 N.N.BAGCHI RD Titagarh Anandaprasad Banerjee Road, Rathtala, 22 7 Katadanga, South A B Road, Singapara, Bhatpara Bhatpara 23 12 Kelabagan Bakar Mahalla, Bhatpara Bhatpara 24 Bhatpara 13 KANKINARA Bhatpara 25 17 Puranitala, Jagaddal Jagaddal 26 19 Rajpukurpath, Authpur, Jagaddal Jagaddal 27 27 Gurdaha natun pally, Shyamnagar Bhatpara 28 33 UTTAR PANPUR, BHATPARA, JAGADDAL Bhatpara Apollo Nursing Home/ Hostel, Gopalpur, Bat- 29 3 Narayanpur tala, Narayanpur Rajarhat, Nutanpara, Pragati Sangha, PO 30 4 Narayanpur Dhapa NISHIKANAN,TEGHARIA,DHALIPARA, 6 (part) CHARNOCK HOSTEL, -
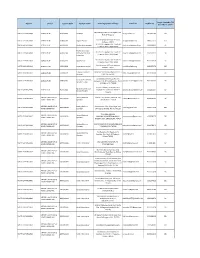
District Sector Course Code Course Name Training Center Address E-Mail ID Mobile No 31St March 2019
Target Available Till District Sector Course Code Course Name Training Center Address E-mail ID Mobile No 31st March 2019 Mama Bhagina Bus stand, Bagdah, Dist- NORTH 24 PARGANAS AGRICULTURE AGR/Q0801 Gardener [email protected] 9874062186 570 Norh 24 Parganas Prafulla Kanan, VIP Road, Kestopur, NORTH 24 PARGANAS AGRICULTURE AGR/Q1201 Organic Grower [email protected] 9830037376 254 Kolkata - 700101 Vill +PO+PS : Bagdah, Dist : North 24 NORTH 24 PARGANAS AGRICULTURE AGR/Q4303 Poultry farm manager [email protected] 9434102419 30 Parganas, State : West Bengal Poultry feed, food Vill +PO+PS : Bagdah, Dist : North 24 NORTH 24 PARGANAS AGRICULTURE AGR/Q4305 safety and labelling [email protected] 9434102419 30 Parganas, State : West Bengal supervisor Vill +PO+PS : Bagdah, Dist : North 24 NORTH 24 PARGANAS AGRICULTURE AGR/Q4501 Goat Farmer [email protected] 9434102419 30 Parganas, State : West Bengal Prafulla Kanan, VIP Road, Kestopur, NORTH 24 PARGANAS AGRICULTURE AGR/Q4904 Aqua culture worker [email protected] 9830037376 200 Kolkata - 700101 Fisheries Extension BARASAT, Stn. Rd, Near Platform no1, 7 NORTH 24 PARGANAS AGRICULTURE AGR/Q5107 [email protected] 9331040169 90 Associate K.N.C. Rd., Kol-124 Gurukul Edutech Duttapukur,Vill : Agriculture Extension NORTH 24 PARGANAS AGRICULTURE AGR/Q7601 Gangapur, P.O. & P.S. Duttapukur, North [email protected] 9051352337 30 Service Provider 24 Parganas Pin 743248 Gurukul Edutech, Haroa,Vill & P.O. Agriculture Extension NORTH 24 PARGANAS AGRICULTURE AGR/Q7601 Gopalpur,P.S. Haroa Dist. North 24 [email protected] 9051352337 60 Service Provider Parganas APPAREL, MADE-UPS & Sewing Machine Plot No - 3B, Block-LA, Sector-III, Salt NORTH 24 PARGANAS AMH/Q0301 [email protected] 9679669547 50 HOME FURNISHING Operator Lake, Kolkata - 700098 APPAREL, MADE-UPS & Sewing Machine Jhautalamore, Post. -
![Containment Zones in North 24 Pgs [ As on 06/05/2020 ] Without Positives](https://docslib.b-cdn.net/cover/5302/containment-zones-in-north-24-pgs-as-on-06-05-2020-without-positives-3205302.webp)
Containment Zones in North 24 Pgs [ As on 06/05/2020 ] Without Positives
Containment Zones in North 24 Pgs [ as on 06/05/2020 ] without positives SL Muni/Block Ward/GP Locality Police Station No N. N. Road, South Dumdum 1 23 Dum Dum Municipality 2 30 Lake Town Block A, Block B Kolkata 89 Lake Town 33(part) 3 34(part), Dakshindari road, Laketown. Lake Town 35 (part) 4 Gorakshabasi Road. Nagerbazar Dum Dum 25 5 Kajipara,Kabi Nabin Sen Rd Dum Dum South Dum Dum 6 21 Nayapati Road Dum Dum Diamond city, Jossore Road 7 27 Dum Dum 8 28 Dum Dum park Lake Town 9 29 Bangur, Laketown Lake Town 10 16 Roy Mallickpara Colony, Dum Dum Dum Dum Rastraguru Avenue, Ramghar 11 8 Dum Dum Colony,NAGAR BAZAR 12 26 Kabi Bharat Chandra Road Dum Dum 13 25 Jogendra Basak Road, Baranagar Baranagar 14 29 Nanda Kumar Road, Baranagar Baranagar 15 Baranagar 13 Gopal Lal Thakur Road, Baranagar Baranagar 16 17 Nabin Chandra Das Road Baranagar 17 4 Kumarpara Lane,Jhilbagan, Kol-35 Baranagar 18 17 Rathtala, Kamarhati Belghoria Club Town Heights, Belgharia, 19 16 Belghoria Rathtala Jaganta Pally, Deshapriya Nagar, 20 25 Belghoria Nandan Kanan, Belghoria 21 26 Rani Park, Belghoria Belghoria 22 Kamarhati 18 Sarat Pally, Belghoria Belghoria 23 1 Banerjee Bagan Kamarhati Belghoria OLD NIMTA RD, Thakurdas 24 21 Banerjee Road, KP Ghoshal Road, Belghoria Belghoria 25 13 Kumud Ghosal Rd., Ariadaha Belghoria 26 4 CHARU MARKET Belghoria 27 2 TOOTH BAGAN Belghoria Airport Gate No. 1,East kamalpur, 28 11 Dum Dum Dum Dum RBC Road, Dum Dum Central Jail 29 20 Dum Dum Mor 30 4 Dum Dum MALANCHA,2.5 NO AIRPORT GATE Dum Dum Swami Swarupananda Sarani, Kol- 31 6 Airport 79 INCINERATOR RD,DUMDUM 32 17 Dum Dum CANTT S P MUKHERJEE RD, DUMDUM 33 19 Dum Dum CANTT 34 19 Birati, Khalisakota Pally Dum Dum 35 36 10 Kabi Satyan dutta road Nimta 37 12 Birati, Nimta, Pathanpur Nimta North Dum Dum 38 21 Srinagar,Durganagar, Birati,Kol-51 Nimta Parts of Purba Alipur (near Gangulybari), 39 Nimta 27, 28, 33 Nimta BL No. -

1ST DROP STOPPAGE TIME Total APONALOY Bus No
ROUTE NO. 01 7713 DROP 1ST DROP STOPPAGE TIME Total APONALOY Bus No. 01 13:42 7 FORTUNE CITY Bus No. 01 13:46 2 MADHYAMGRAM CHOWMATHA Bus No. 01 13:50 9 MADHYAMGRAM SRACHI Bus No. 01 13:52 4 SISIR KUNJA Bus No. 01 13:53 5 DAKBANGLA MORE Bus No. 01 13:56 2 BARASAT 11 NO. RAIL GATE Bus No. 01 13:58 2 HELA BATTALA MORE Bus No. 01 14:00 5 36 ROUTE NO. 02 4545 DROP STOPPAGE TIME Total TETULTALA BUS STOP Bus No. 02 13:45 2 BELGACHHIA MILK COLONY Bus No. 02 13:48 4 BELGACHHIA TRAM DEPO. Bus No. 02 13:51 2 TALA POLICE STATION Bus No. 02 13:52 1 TALA LADIES PARK Bus No. 02 13:53 2 TALA PARK Bus No. 02 13:54 1 PAIKPARA 2 NO. BUS STAND Bus No. 02 13:55 1 PAIKPARA BT ROAD XING Bus No. 02 13:56 2 RABINDRA BHARTI UNIVERSITY Bus No. 02 14:02 4 SINTHEE MORE Bus No. 02 14:04 3 B.T. TOBIN RD XING Bus No. 02 14:05 1 BON HOOGLY Bus No. 02 14:07 2 DUNLOP Bus No. 02 14:08 4 RATHTALA Bus No. 02 14:11 3 SODEPUR (PEERLESS NAGAR) Bus No. 02 14:15 4 36 ROUTE NO. 03 1059 DROP STOPPAGE TIME Total AIRPORT 1 NO. KALIMANDIR Bus No. 03 13:28 1 NATIONAL OPTICAL Bus No. 03 13:30 1 JK GARDEN APT. Bus No. 03 13:31 1 SIDDHESHWARI KALIBARI Bus No. 03 13:32 3 KRISHI GOPAL MORE Bus No. -

List of Covid Vaccination Session Sites on 28-06-2021
List of Covid Vaccination Session Sites on 28-06-2021 Type of Type of Vaccine CVC/WPCVC Sl. No. District Name of Block / ULB Name of CVC / WPCVC CVC (Govt. (Covishield / Covaxin / ID or Pvt.) Sputnik V) 1 Birbhum Nanoor CVC Nanoor RH Govt. Covishield 2 Birbhum Nanoor CVC KHALA HWC Govt. Covishield 3 Birbhum Nanoor CVC PAKURHANS Sc Govt. Covishield 4 Birbhum Nanoor CVC BRAHMANKHANDA HWC Govt. Covishield 5 Birbhum Nanoor CVC NANOOR SC Govt. Covishield 6 Birbhum Bolpur WPCVC Bolpur BPHC 18-44 Covaxin Govt. Covaxin 7 Birbhum Suri-II WPCVC BANSRA HWC STATE 18 TO 45 Govt. Covishield 8 Birbhum Sainthia CVC Sainthia RH Govt. Covishield 9 Birbhum Suri-I CVC Barachaturi SC Govt. Covishield 10 Birbhum Rajnagar WPCVC RAJNAGAR TRANSPORT REPORTER Govt. Covishield 11 Birbhum Suri-I CVC Kalipur Govt. Covishield 12 Birbhum Sainthia WPCVC Sainthia GP Hawker above 45 Govt. Covishield 13 Birbhum Sainthia WPCVC Sainthia Govt. Emp. Below 45 Govt. Covishield 14 Birbhum Suri Urban WPCVC SURI DH COVX 18-44 Govt. Covaxin 15 Birbhum Suri Urban WPCVC Medica Nirmala Devi NH Pvt. Covishield 16 Birbhum Suri-I CVC Tilpara HWC Govt. Covishield 17 Birbhum Sainthia CVC Rajnagar RH Govt. Covishield 18 Birbhum Sainthia CVC Sainthia RH COV Govt. Covishield 19 Birbhum Suri-II CVC Bansra Hwc Govt. Covishield 20 Birbhum Sainthia WPCVC Sainthia GP Hawker below 45 Govt. Covishield 21 Birbhum Suri Urban WPCVC Suri DH Transport Govt. Covishield 22 Birbhum Sainthia WPCVC Sainthia Govt. Emp. Above 45 Govt. Covishield 23 Birbhum Nanoor CVC UCHKARAN S C Govt. Covishield 24 Birbhum Sainthia WPCVC Sainthia Covax 18-44 Govt.