A Reappraisal of the Systematic Affinities of Socotran, Arabian And
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Island Biology Island Biology
IIssllaanndd bbiioollooggyy Allan Sørensen Allan Timmermann, Ana Maria Martín González Camilla Hansen Camille Kruch Dorte Jensen Eva Grøndahl, Franziska Petra Popko, Grete Fogtmann Jensen, Gudny Asgeirsdottir, Hubertus Heinicke, Jan Nikkelborg, Janne Thirstrup, Karin T. Clausen, Karina Mikkelsen, Katrine Meisner, Kent Olsen, Kristina Boros, Linn Kathrin Øverland, Lucía de la Guardia, Marie S. Hoelgaard, Melissa Wetter Mikkel Sørensen, Morten Ravn Knudsen, Pedro Finamore, Petr Klimes, Rasmus Højer Jensen, Tenna Boye Tine Biedenweg AARHUS UNIVERSITY 2005/ESSAYS IN EVOLUTIONARY ECOLOGY Teachers: Bodil K. Ehlers, Tanja Ingversen, Dave Parker, MIchael Warrer Larsen, Yoko L. Dupont & Jens M. Olesen 1 C o n t e n t s Atlantic Ocean Islands Faroe Islands Kent Olsen 4 Shetland Islands Janne Thirstrup 10 Svalbard Linn Kathrin Øverland 14 Greenland Eva Grøndahl 18 Azores Tenna Boye 22 St. Helena Pedro Finamore 25 Falkland Islands Kristina Boros 29 Cape Verde Islands Allan Sørensen 32 Tristan da Cunha Rasmus Højer Jensen 36 Mediterranean Islands Corsica Camille Kruch 39 Cyprus Tine Biedenweg 42 Indian Ocean Islands Socotra Mikkel Sørensen 47 Zanzibar Karina Mikkelsen 50 Maldives Allan Timmermann 54 Krakatau Camilla Hansen 57 Bali and Lombok Grete Fogtmann Jensen 61 Pacific Islands New Guinea Lucía de la Guardia 66 2 Solomon Islands Karin T. Clausen 70 New Caledonia Franziska Petra Popko 74 Samoa Morten Ravn Knudsen 77 Tasmania Jan Nikkelborg 81 Fiji Melissa Wetter 84 New Zealand Marie S. Hoelgaard 87 Pitcairn Katrine Meisner 91 Juan Fernandéz Islands Gudny Asgeirsdottir 95 Hawaiian Islands Petr Klimes 97 Galápagos Islands Dorthe Jensen 102 Caribbean Islands Cuba Hubertus Heinicke 107 Dominica Ana Maria Martin Gonzalez 110 Essay localities 3 The Faroe Islands Kent Olsen Introduction The Faroe Islands is a treeless archipelago situated in the heart of the warm North Atlantic Current on the Wyville Thompson Ridge between 61°20’ and 62°24’ N and between 6°15’ and 7°41’ W. -

Tc & Forward & Owls-I-IX
USDA Forest Service 1997 General Technical Report NC-190 Biology and Conservation of Owls of the Northern Hemisphere Second International Symposium February 5-9, 1997 Winnipeg, Manitoba, Canada Editors: James R. Duncan, Zoologist, Manitoba Conservation Data Centre Wildlife Branch, Manitoba Department of Natural Resources Box 24, 200 Saulteaux Crescent Winnipeg, MB CANADA R3J 3W3 <[email protected]> David H. Johnson, Wildlife Ecologist Washington Department of Fish and Wildlife 600 Capitol Way North Olympia, WA, USA 98501-1091 <[email protected]> Thomas H. Nicholls, retired formerly Project Leader and Research Plant Pathologist and Wildlife Biologist USDA Forest Service, North Central Forest Experiment Station 1992 Folwell Avenue St. Paul, MN, USA 55108-6148 <[email protected]> I 2nd Owl Symposium SPONSORS: (Listing of all symposium and publication sponsors, e.g., those donating $$) 1987 International Owl Symposium Fund; Jack Israel Schrieber Memorial Trust c/o Zoological Society of Manitoba; Lady Grayl Fund; Manitoba Hydro; Manitoba Natural Resources; Manitoba Naturalists Society; Manitoba Critical Wildlife Habitat Program; Metro Propane Ltd.; Pine Falls Paper Company; Raptor Research Foundation; Raptor Education Group, Inc.; Raptor Research Center of Boise State University, Boise, Idaho; Repap Manitoba; Canadian Wildlife Service, Environment Canada; USDI Bureau of Land Management; USDI Fish and Wildlife Service; USDA Forest Service, including the North Central Forest Experiment Station; Washington Department of Fish and Wildlife; The Wildlife Society - Washington Chapter; Wildlife Habitat Canada; Robert Bateman; Lawrence Blus; Nancy Claflin; Richard Clark; James Duncan; Bob Gehlert; Marge Gibson; Mary Houston; Stuart Houston; Edgar Jones; Katherine McKeever; Robert Nero; Glenn Proudfoot; Catherine Rich; Spencer Sealy; Mark Sobchuk; Tom Sproat; Peter Stacey; and Catherine Thexton. -

Winter Thermoregulation in Free-Ranging Pearl-Spotted Owlets and African Scops-Owls
149 Do Owls Use Torpor? Winter Thermoregulation in Free-Ranging Pearl-Spotted Owlets and African Scops-Owls Ben Smit* 6,500 g, and in a much wider variety of ecological contexts Andrew E. McKechnie† than previously thought (Ko¨rtner et al. 2000; McKechnie and DST/NRF Centre of Excellence at the Percy FitzPatrick Lovegrove 2002). Within the Coraciiformes, for instance, het- Institute, School of Animal, Plant and Environmental erothermy occurs in both the smallest and largest representa- Sciences, University of the Witwatersrand, Private Bag 3, tives, namely, the 5-g Puerto Rican tody Todus mexicanus and Wits 2050, South Africa the 360-g laughing kookaburra Dacelo novaeguineae (Merola- Zwartjes and Ligon 2000; Cooper et al. 2008). Accepted 3/6/2009; Electronically Published 11/24/2009 Torpor, often defined as a reduction in rest-phase Tb below 30ЊC (Hudson 1978; Reinertsen 1996; Schleucher 2001; re- viewed by Barclay et al. 2001), appears to be particularly im- portant in offsetting the energetic costs of thermoregulation in ABSTRACT birds whose food resources exhibit large spatial or temporal Numerous avian taxa use torpor, which involves pronounced fluctuations in availability, for example, frugivores (Coliidae reductions in body temperature (Tb) to below normothermic and Columbidae), nectarivores (Trochilidae and Nectariniidae), levels. However, the occurrence of this phenomenon in owls and insectivores (Todidae, Apodidae, Caprimulgidae, and Hi- (Strigidae) remains largely unknown. We investigated winter rundinidae; McKechnie and Lovegrove 2002; Schleucher 2004). patterns of thermoregulation in the crepuscular 80-g pearl- However, to better understand the adaptive value and evolution spotted owlet Glaucidium perlatum and the strictly nocturnal of avian torpor, a more thorough assessment of the phylogenetic 61-g African scops-owl Otus senegalensis by obtaining telemetric distribution of this trait is needed (McKechnie and Lovegrove measurements of skin temperature (Tskin) from free-ranging 2002; Schleucher 2004). -

Raptors in the East African Tropics and Western Indian Ocean Islands: State of Ecological Knowledge and Conservation Status
j. RaptorRes. 32(1):28-39 ¸ 1998 The Raptor ResearchFoundation, Inc. RAPTORS IN THE EAST AFRICAN TROPICS AND WESTERN INDIAN OCEAN ISLANDS: STATE OF ECOLOGICAL KNOWLEDGE AND CONSERVATION STATUS MUNIR VIRANI 1 AND RICHARD T. WATSON ThePeregrine Fund, Inc., 566 WestFlying Hawk Lane, Boise,1D 83709 U.S.A. ABSTRACT.--Fromour reviewof articlespublished on diurnal and nocturnal birds of prey occurringin Africa and the western Indian Ocean islands,we found most of the information on their breeding biology comesfrom subtropicalsouthern Africa. The number of published papers from the eastAfrican tropics declined after 1980 while those from subtropicalsouthern Africa increased.Based on our KnoM- edge Rating Scale (KRS), only 6.3% of breeding raptorsin the eastAfrican tropicsand 13.6% of the raptorsof the Indian Ocean islandscan be consideredWell Known,while the majority,60.8% in main- land east Africa and 72.7% in the Indian Ocean islands, are rated Unknown. Human-caused habitat alteration resultingfrom overgrazingby livestockand impactsof cultivationare the main threatsfacing raptors in the east African tropics, while clearing of foreststhrough slash-and-burnmethods is most important in the Indian Ocean islands.We describeconservation recommendations, list priorityspecies for study,and list areasof ecologicalunderstanding that need to be improved. I•y WORDS: Conservation;east Africa; ecology; western Indian Ocean;islands; priorities; raptors; research. Aves rapacesen los tropicos del este de Africa yen islasal oeste del Oc•ano Indico: estado del cono- cimiento eco16gicoy de su conservacitn RESUMEN.--Denuestra recopilacitn de articulospublicados sobre aves rapaces diurnas y nocturnasque se encuentran en Africa yen las islasal oeste del Octano Indico, encontramosque la mayoriade la informaci6n sobre aves rapacesresidentes se origina en la regi6n subtropical del sur de Africa. -

Strigiformes) and Lesser Nighthawks (Chodeiles Acutipennis
UNIVERSITY OF CALIFORNIA RIVERSIDE The Evolution of Quiet Flight in Owls (Strigiformes) and Lesser Nighthawks (Chodeiles acutipennis) A Dissertation submitted in partial satisfaction of the requirements for the degree of Doctor of Philosophy in Evolution, Ecology, and Organismal Biology by Krista Le Piane December 2020 Dissertation Committee: Dr. Christopher J. Clark, Chairperson Dr. Erin Wilson Rankin Dr. Khaleel A. Razak Copyright by Krista Le Piane 2020 The Dissertation of Krista Le Piane is approved: Committee Chairperson University of California, Riverside ACKNOWLEDGEMENTS I thank my Oral Exam Committee: Dr. Khaleel A. Razak (chairperson), Dr. Erin Wilson Rankin, Dr. Mark Springer, Dr. Jesse Barber, and Dr. Scott Curie. Thank you to my Dissertation Committee: Dr. Christopher J. Clark (chairperson), Dr. Erin Wilson Rankin, and Dr. Khaleel A. Razak for their encouragement and help with this dissertation. Thank you to my lab mates, past and present: Dr. Sean Wilcox, Dr. Katie Johnson, Ayala Berger, David Rankin, Dr. Nadje Najar, Elisa Henderson, Dr. Brian Meyers Dr. Jenny Hazelhurst, Emily Mistick, Lori Liu, and Lilly Hollingsworth for their friendship and support. I thank the Natural History Museum of Los Angeles County (LACM), the California Academy of Sciences (CAS), Museum of Vertebrate Zoology (MVZ) at UC Berkeley, the American Museum of Natural History (ANMH), and the Natural History Museum (NHM) in Tring for access to specimens used in Chapter 1. I would especially like to thank Kimball Garrett and Allison Shultz for help at LACM. I also thank Ben Williams, Richard Jackson, and Reddit user NorthernJoey for permission to use their photos in Chapter 1. Jessica Tingle contributed R code and advice to Chapter 1 and I would like to thank her for her help. -

Habitat Selection by Terrestrial Birds on Pemba Island (Tanzania), with Particular Reference to Six Endemic Taxa
Biological Conservation 95 (2000) 259±267 www.elsevier.com/locate/biocon Habitat selection by terrestrial birds on Pemba Island (Tanzania), with particular reference to six endemic taxa Paulo Catry a,b,*, Richard Mellanby c, K. Ali. Suleiman d, K. Haji Salim d, M. Hughes c, M. McKean c, N. Anderson c, G. Constant c, V. Heany c, G. Martin c, M. Armitage c, M. Wilson c aUnidade de InvestigacËaÄo em Eco-Etologia, ISPA, R. Jardim do Tabaco 44, 1100 Lisboa, Portugal bEdward Grey Institute of Ornithology, Zoology Department, University of Oxford, UK cProject Pemba. Glasgow University Expedition 1998. IBLS, University of Glasgow, Glasgow G12 8QQ, UK dCommission for Natural Resources, PO Box 283, Wete-Pemba, Tanzania Received 16 September 1999; received in revised form 9 February 2000; accepted 8 March 2000 Abstract An important proportion of the world's biodiversity is found on oceanic islands. Island endemics frequently have small popula- tions and are known to be sensitive to habitat and community changes, making them prone to extinction. In this paper, we assess the habitat distribution of the terrestrial birds of Pemba, an oceanic island that has been classi®ed has an ``Endemic Bird Area''. Most of Pemba has been profoundly altered by human activities and only small patches of natural vegetation remain. However, we found that the six endemic birds (four species and two sub-species) have colonised several of the man-made habitats, including clove plantations and farmland, and remain widespread. Species richness was not reduced in these heavily managed areas when compared to the remnants of tropical forest. -

SOUTHERN INDIA and SRI LANKA
Sri Lanka Woodpigeon (all photos by D.Farrow unless otherwise stated) SOUTHERN INDIA and SRI LANKA (WITH ANDAMANS ISLANDS EXTENSION) 25 OCTOBER – 19 NOVEMBER 2016 LEADER: DAVE FARROW This years’ tour to Southern India and Sri Lanka was once again a very successful and enjoyable affair. A wonderful suite of endemics were seen, beginning with our extension to the Andaman Islands where we were able to find 20 of the 21 endemics, with Andaman Scops and Walden’s Scops Owls, Andaman and Hume’s Hawk Owls leading the way, Andaman Woodpigeon and Andaman Cuckoo Dove, good looks at 1 BirdQuest Tour Report: South India and Sri Lanka 2016 www.birdquest-tours.com Andaman Crake, plus all the others with the title ‘Andaman’ (with the exception of the Barn Owl) and a rich suite of other birds such as Ruddy Kingfisher, Oriental Pratincole, Long-toed Stint, Long-tailed Parakeets and Mangrove Whistler. In Southern India we birded our way from the Nilgiri Hills to the lowland forest of Kerala finding Painted and Jungle Bush Quail, Jungle Nightjar, White-naped and Heart-spotted Woodpeckers, Malabar Flameback, Malabar Trogons, Malabar Barbet, Blue-winged Parakeet, Grey-fronted Green Pigeons, Nilgiri Woodpigeon, Indian Pitta (with ten seen on the tour overall), Jerdon's Bushlarks, Malabar Larks, Malabar Woodshrike and Malabar Whistling Thrush, Black-headed Cuckooshrike, Black-and- Orange, Nilgiri, Brown-breasted and Rusty-tailed Flycatchers, Nilgiri and White-bellied Blue Robin, Black- chinned and Kerala Laughingthrushes, Dark-fronted Babblers, Indian Rufous Babblers, Western Crowned Warbler, Indian Yellow Tit, Indian Blackbird, Hill Swallow, Nilgiri Pipit, White-bellied Minivet, the scarce Yellow-throated and Grey-headed Bulbuls, Flame-throated and Yellow-browed Bulbuls, Nilgiri Flowerpecker, Loten's Sunbird, Black-throated Munias and the stunning endemic White-bellied Treepie. -

Familiar Chat
phone 3190540 email [email protected] Familiar Chat N e w s l e t t e r o f B i r d L i f e B o t s w a n a About Bird Walks WHO? If you are interested in learning about birds, you are welcome! WHAT? We walk slowly for a couple of kilometres and look for birds. You need binoculars, a bird ID book is useful. A vehicle which can deal with bumpy, overgrown tracks is useful, but not essential. However - don’t set off without folding chairs and the makings of a picnic. WHEN and WHERE? The 1st Sunday and the 3rd Saturday of the month at 6.30am meeting on the Molapo Crossing Car Park. The Sunday walks are permanent fixtures as demand is Researchers evaluated 1,991 participants from the Walking high. The Saturday programme, for beginners only, is a for Health program in England, which helps facilitate nearly recent addition and depends on the level of interest. Check 3,000 weekly walks and draws more than 70,000 regular the website for details. walkers a year. WHY? “Walking is an inexpensive, low risk and accessible “Given the increase in mental ill health and physical form of exercise and it turns inactivity in the developed world, we are constantly out that combined with nature exploring new, accessible ways to help people improve their and group settings, it may be a long term quality of life and well-being,” Grey Crowned Crane – endangered very powerful, under-utilized “Group walks in local natural environments may make a through habitat destruction stress buster. -
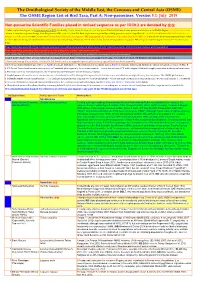
ORL 5.1 Non-Passerines Final Draft01a.Xlsx
The Ornithological Society of the Middle East, the Caucasus and Central Asia (OSME) The OSME Region List of Bird Taxa, Part A: Non-passerines. Version 5.1: July 2019 Non-passerine Scientific Families placed in revised sequence as per IOC9.2 are denoted by ֍֍ A fuller explanation is given in Explanation of the ORL, but briefly, Bright green shading of a row (eg Syrian Ostrich) indicates former presence of a taxon in the OSME Region. Light gold shading in column A indicates sequence change from the previous ORL issue. For taxa that have unproven and probably unlikely presence, see the Hypothetical List. Red font indicates added information since the previous ORL version or the Conservation Threat Status (Critically Endangered = CE, Endangered = E, Vulnerable = V and Data Deficient = DD only). Not all synonyms have been examined. Serial numbers (SN) are merely an administrative convenience and may change. Please do not cite them in any formal correspondence or papers. NB: Compass cardinals (eg N = north, SE = southeast) are used. Rows shaded thus and with yellow text denote summaries of problem taxon groups in which some closely-related taxa may be of indeterminate status or are being studied. Rows shaded thus and with yellow text indicate recent or data-driven major conservation concerns. Rows shaded thus and with white text contain additional explanatory information on problem taxon groups as and when necessary. English names shaded thus are taxa on BirdLife Tracking Database, http://seabirdtracking.org/mapper/index.php. Nos tracked are small. NB BirdLife still lump many seabird taxa. A broad dark orange line, as below, indicates the last taxon in a new or suggested species split, or where sspp are best considered separately. -

EUROPEAN BIRDS of CONSERVATION CONCERN Populations, Trends and National Responsibilities
EUROPEAN BIRDS OF CONSERVATION CONCERN Populations, trends and national responsibilities COMPILED BY ANNA STANEVA AND IAN BURFIELD WITH SPONSORSHIP FROM CONTENTS Introduction 4 86 ITALY References 9 89 KOSOVO ALBANIA 10 92 LATVIA ANDORRA 14 95 LIECHTENSTEIN ARMENIA 16 97 LITHUANIA AUSTRIA 19 100 LUXEMBOURG AZERBAIJAN 22 102 MACEDONIA BELARUS 26 105 MALTA BELGIUM 29 107 MOLDOVA BOSNIA AND HERZEGOVINA 32 110 MONTENEGRO BULGARIA 35 113 NETHERLANDS CROATIA 39 116 NORWAY CYPRUS 42 119 POLAND CZECH REPUBLIC 45 122 PORTUGAL DENMARK 48 125 ROMANIA ESTONIA 51 128 RUSSIA BirdLife Europe and Central Asia is a partnership of 48 national conservation organisations and a leader in bird conservation. Our unique local to global FAROE ISLANDS DENMARK 54 132 SERBIA approach enables us to deliver high impact and long term conservation for the beneit of nature and people. BirdLife Europe and Central Asia is one of FINLAND 56 135 SLOVAKIA the six regional secretariats that compose BirdLife International. Based in Brus- sels, it supports the European and Central Asian Partnership and is present FRANCE 60 138 SLOVENIA in 47 countries including all EU Member States. With more than 4,100 staf in Europe, two million members and tens of thousands of skilled volunteers, GEORGIA 64 141 SPAIN BirdLife Europe and Central Asia, together with its national partners, owns or manages more than 6,000 nature sites totaling 320,000 hectares. GERMANY 67 145 SWEDEN GIBRALTAR UNITED KINGDOM 71 148 SWITZERLAND GREECE 72 151 TURKEY GREENLAND DENMARK 76 155 UKRAINE HUNGARY 78 159 UNITED KINGDOM ICELAND 81 162 European population sizes and trends STICHTING BIRDLIFE EUROPE GRATEFULLY ACKNOWLEDGES FINANCIAL SUPPORT FROM THE EUROPEAN COMMISSION. -

Nr. C 253/18 Amtsblatt Der Europäischen Gemeinschaften 10
Nr. C 253/18 Amtsblatt der Europäischen Gemeinschaften 10. 10. 86 PROTOCOL CONCERNING PROTECTED AREAS AND WILD FAUNA AND FLORA IN THE EASTERN AFRICAN REGION THE CONTRACTING PARTIES TO THE PRESENT PROTOCOL, Being parties to the Convention for the protection, management and development of the marine and coastal environment of the Eastern African region, done at Nairobi on 21 June 1985, Conscious of the danger from increasing human activities which are threatening the environment of the Eastern African region, Recognizing that natural resources constitute a heritage of scientific, cultural, educational, recreational and economic value that needs to be effectively protected, Stressing the importance of protecting and, as appropriate, improving the State of the wild fauna and flora and natural habitats of the Eastern African region among other means by the establishment of specially protected areas in the marine and coastal environment, Desirous of establishing close Cooperation among themselves in order to achieve that objective, HAVE AGREED AS FOLLOWS: Article 1 Article 3 Definitions Protection of wild flora For the purposes of this Protocol: The Contracting Parties shall take all appropriate (a) 'Eastern African region' means the Convention area measures to ensure the protection of the wild flora as defined in paragraph (a) of Article 2 of the species specified in Annex I. To this end, each Convention. It shall also include the coastal areas of Contracting Party shall, as appropriate, prohibit activities the Contracting Parties and their internal waters having adverse effects on the habitats of such species, as related to the marine and coastal environment; well as the uncontrolled picking, collecting, cutting or uprooting of such species. -

Namibia's Etosha Pan & Skeleton Coast
Namibia's Etosha Pan & Skeleton Coast Naturetrek Tour Report 30 October - 15 November 2015 Black Rhinoceros Elephant Family Flamingoes at Walvis Bay The desert Report compiled by Rob Mileto Images courtesy of Ingrid William Naturetrek Mingledown Barn Wolf's Lane Chawton Alton Hampshire GU34 3HJ UK T: +44 (0)1962 733051 E: [email protected] W: www.naturetrek.co.uk Tour Report Namibia's Etosha Pan & Skeleton Coast Tour Participants: Rob Mileto, Festus Mbinga & Franco Morao (leaders) and 12 Naturetrek clients Day 1 Friday 30th October London Heathrow to Johannesburg We all met up, mostly at the gate, for an uneventful overnight flight to Johannesburg in our double-decker plane Day 2 Saturday 31st October Johannesburg to Namib Grens Farm (via Windhoek) Weather: hot and sunny. The bleary but keen-eyed spotted our first southern African bird, a Rock Martin, from the Johannesburg airport terminal building. After a welcome coffee or two, a further short flight over the Kalahari brought us to Windhoek. Here we met out local guides, Festus and Franco, and were soon aboard our extended Land Rovers that were to be our transport and ‘hides’ for the next two weeks. Then we were off. After passing through Windhoek, we were soon out in the wilds and spotting lots of new birds and mammals like Chacma Baboon, Springbok, Cape Starling, Southern Yellow-billed Hornbill, White- backed Mousebird, Pale Chanting Goshawk and Ostrich. All these distractions meant that we arrived at Namib Grens after dark. The bungalows here are literally built around granite boulders which form some of the walls, and after a hearty farm dinner we retired to our beds amongst the rocks – one complete with a Rock Hyrax stuck in the bath! Day 3 Sunday 1st November Namib Grens to Kulala Weather: hot and sunny.