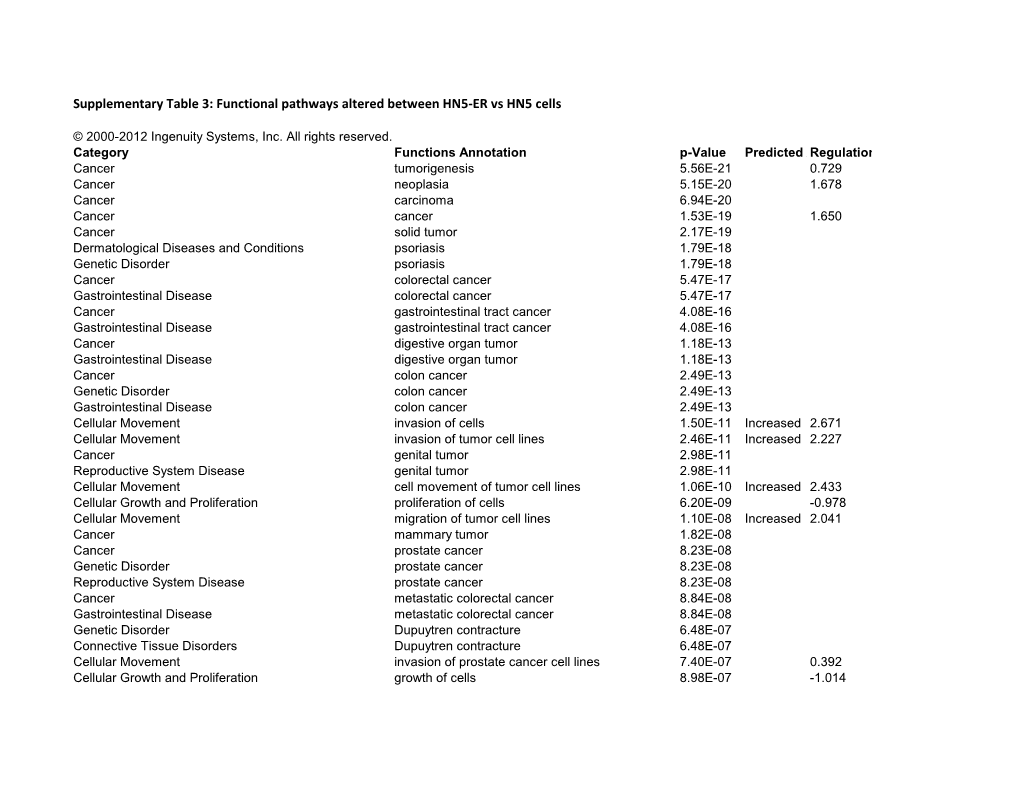
Supplementary Table 3: Functional Pathways Altered Between HN5-ER Vs HN5 Cells
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 T + is online at: average * The Journal of Immunology , 34 of which you can access for free at: 2016; 197:1477-1488; Prepublished online 1 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600589 http://www.jimmunol.org/content/197/4/1477 Molecular Profile of Tumor-Specific CD8 Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh, Sonia M. Leach, Brandon L. Moore, Tullia C. Bruno, Jonathan D. Buhrman and Jill E. Slansky J Immunol cites 95 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2016/07/01/jimmunol.160058 9.DCSupplemental This article http://www.jimmunol.org/content/197/4/1477.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. The Journal of Immunology Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. -

Epidermal Fatty Acid-Binding Protein 5 (FABP5) Involvement in Alpha-Synuclein-Induced Mitochondrial Injury Under Oxidative Stress
biomedicines Article Epidermal Fatty Acid-Binding Protein 5 (FABP5) Involvement in Alpha-Synuclein-Induced Mitochondrial Injury under Oxidative Stress Yifei Wang , Yasuharu Shinoda , An Cheng , Ichiro Kawahata and Kohji Fukunaga * Department of Pharmacology, Graduate School of Pharmaceutical Sciences, Tohoku University, 6–3 Aramaki-Aoba, Aoba-ku, Sendai 980-8578, Japan; [email protected] (Y.W.); [email protected] (Y.S.); [email protected] (A.C.); [email protected] (I.K.) * Correspondence: [email protected]; Tel.: +81-22-795-6836 Abstract: The accumulation of α-synuclein (αSyn) has been implicated as a causal factor in the pathogenesis of Parkinson’s disease (PD). There is growing evidence that supports mitochondrial dysfunction as a potential primary cause of dopaminergic neuronal death in PD. Here, we focused on reciprocal interactions between αSyn aggregation and mitochondrial injury induced by oxidative stress. We further investigated whether epidermal fatty acid-binding protein 5 (FABP5) is related to αSyn oligomerization/aggregation and subsequent disturbances in mitochondrial function in neuronal cells. In the presence of rotenone, a mitochondrial respiratory chain complex I inhibitor, co- overexpression of FABP5 with αSyn significantly decreased the viability of Neuro-2A cells compared to that of αSyn alone. Under these conditions, FABP5 co-localized with αSyn in the mitochondria, thereby reducing mitochondrial membrane potential. Furthermore, we confirmed that pharmacologi- cal inhibition of FABP5 by its ligand prevented αSyn accumulation in mitochondria, which led to cell death rescue. These results suggested that FABP5 is crucial for mitochondrial dysfunction related to Citation: Wang, Y.; Shinoda, Y.; αSyn oligomerization/aggregation in the mitochondria induced by oxidative stress in neurons. -

Enrichment and Characterization of Two Subgroups of Committed Osteogenic Cells in the Mouse Endosteal Bone Marrow with Expressio
ne Marro Bo w f R o e l s a e n a r Chang CF et al., J Bone Marrow Res 2014, 2:2 r c u h o J Journal of Bone Marrow Research DOI: 10.4172/2329-8820.1000144 ISSN: 2329-8820 Research Article Open Access Enrichment and Characterization of Two Subgroups of Committed Osteogenic Cells in the Mouse Endosteal Bone Marrow with Expression Levels of CD24 Ching-Fang Chang1,2, Ke-Hsun Hsu1,2, Chia-Ning Shen1,2,3, Chung-Leung Li2,3* and Jean Lu1,2** 1Institute of Microbiology and Immunology, National Yang-Ming University, Taipei, Taiwan 2Genomics Research Center, Academia Sinica, Taipei, Taiwan 3Institute of Bioscience and Biotechnology, College of Life Sciences, National Taiwan Ocean University, Keelung, Taiwan Abstract Primary osteogenic cells have been known to reside within the CD45-CD31-Ter119-Sca-1- cell fraction, particularly in the CD51+ subpopulation. However, detailed determination of the frequency of osteogenic cells within this Sca-1- cell population remains yet to be determined. In addition, it is not clear that other cell surface markers can be used to further sub-fractionate this Sca-1-CD51+ osteogenic cell population and to define their developmental stages. In this report, both Sca-1-CD24med and Sca-1- CD24-/lo cells have been shown to be two small subsets of the Sca-1-CD51+ cell fraction. These two cell fractions show subtle difference in the expression level of osteogenic marker genes such as Osx and Opn, and in vitro proliferate rate. All these observations suggest that they may be at different developmental stages of osteogenesis. -

An Amplified Fatty Acid-Binding Protein Gene Cluster In
cancers Review An Amplified Fatty Acid-Binding Protein Gene Cluster in Prostate Cancer: Emerging Roles in Lipid Metabolism and Metastasis Rong-Zong Liu and Roseline Godbout * Department of Oncology, Cross Cancer Institute, University of Alberta, Edmonton, AB T6G 1Z2, Canada; [email protected] * Correspondence: [email protected]; Tel.: +1-780-432-8901 Received: 6 November 2020; Accepted: 16 December 2020; Published: 18 December 2020 Simple Summary: Prostate cancer is the second most common cancer in men. In many cases, prostate cancer grows very slowly and remains confined to the prostate. These localized cancers can usually be cured. However, prostate cancer can also metastasize to other organs of the body, which often results in death of the patient. We found that a cluster of genes involved in accumulation and utilization of fats exists in multiple copies and is expressed at much higher levels in metastatic prostate cancer compared to localized disease. These genes, called fatty acid-binding protein (or FABP) genes, individually and collectively, promote properties associated with prostate cancer metastasis. We propose that levels of these FABP genes may serve as an indicator of prostate cancer aggressiveness, and that inhibiting the action of FABP genes may provide a new approach to prevent and/or treat metastatic prostate cancer. Abstract: Treatment for early stage and localized prostate cancer (PCa) is highly effective. Patient survival, however, drops dramatically upon metastasis due to drug resistance and cancer recurrence. The molecular mechanisms underlying PCa metastasis are complex and remain unclear. It is therefore crucial to decipher the key genetic alterations and relevant molecular pathways driving PCa metastatic progression so that predictive biomarkers and precise therapeutic targets can be developed. -

Single-Cell RNA Sequencing Demonstrates the Molecular and Cellular Reprogramming of Metastatic Lung Adenocarcinoma
ARTICLE https://doi.org/10.1038/s41467-020-16164-1 OPEN Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma Nayoung Kim 1,2,3,13, Hong Kwan Kim4,13, Kyungjong Lee 5,13, Yourae Hong 1,6, Jong Ho Cho4, Jung Won Choi7, Jung-Il Lee7, Yeon-Lim Suh8,BoMiKu9, Hye Hyeon Eum 1,2,3, Soyean Choi 1, Yoon-La Choi6,10,11, Je-Gun Joung1, Woong-Yang Park 1,2,6, Hyun Ae Jung12, Jong-Mu Sun12, Se-Hoon Lee12, ✉ ✉ Jin Seok Ahn12, Keunchil Park12, Myung-Ju Ahn 12 & Hae-Ock Lee 1,2,3,6 1234567890():,; Advanced metastatic cancer poses utmost clinical challenges and may present molecular and cellular features distinct from an early-stage cancer. Herein, we present single-cell tran- scriptome profiling of metastatic lung adenocarcinoma, the most prevalent histological lung cancer type diagnosed at stage IV in over 40% of all cases. From 208,506 cells populating the normal tissues or early to metastatic stage cancer in 44 patients, we identify a cancer cell subtype deviating from the normal differentiation trajectory and dominating the metastatic stage. In all stages, the stromal and immune cell dynamics reveal ontological and functional changes that create a pro-tumoral and immunosuppressive microenvironment. Normal resident myeloid cell populations are gradually replaced with monocyte-derived macrophages and dendritic cells, along with T-cell exhaustion. This extensive single-cell analysis enhances our understanding of molecular and cellular dynamics in metastatic lung cancer and reveals potential diagnostic and therapeutic targets in cancer-microenvironment interactions. 1 Samsung Genome Institute, Samsung Medical Center, Seoul 06351, Korea. -

Profiling Data
Compound Name DiscoveRx Gene Symbol Entrez Gene Percent Compound Symbol Control Concentration (nM) JNK-IN-8 AAK1 AAK1 69 1000 JNK-IN-8 ABL1(E255K)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317I)-nonphosphorylated ABL1 87 1000 JNK-IN-8 ABL1(F317I)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317L)-nonphosphorylated ABL1 65 1000 JNK-IN-8 ABL1(F317L)-phosphorylated ABL1 61 1000 JNK-IN-8 ABL1(H396P)-nonphosphorylated ABL1 42 1000 JNK-IN-8 ABL1(H396P)-phosphorylated ABL1 60 1000 JNK-IN-8 ABL1(M351T)-phosphorylated ABL1 81 1000 JNK-IN-8 ABL1(Q252H)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(Q252H)-phosphorylated ABL1 56 1000 JNK-IN-8 ABL1(T315I)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(T315I)-phosphorylated ABL1 92 1000 JNK-IN-8 ABL1(Y253F)-phosphorylated ABL1 71 1000 JNK-IN-8 ABL1-nonphosphorylated ABL1 97 1000 JNK-IN-8 ABL1-phosphorylated ABL1 100 1000 JNK-IN-8 ABL2 ABL2 97 1000 JNK-IN-8 ACVR1 ACVR1 100 1000 JNK-IN-8 ACVR1B ACVR1B 88 1000 JNK-IN-8 ACVR2A ACVR2A 100 1000 JNK-IN-8 ACVR2B ACVR2B 100 1000 JNK-IN-8 ACVRL1 ACVRL1 96 1000 JNK-IN-8 ADCK3 CABC1 100 1000 JNK-IN-8 ADCK4 ADCK4 93 1000 JNK-IN-8 AKT1 AKT1 100 1000 JNK-IN-8 AKT2 AKT2 100 1000 JNK-IN-8 AKT3 AKT3 100 1000 JNK-IN-8 ALK ALK 85 1000 JNK-IN-8 AMPK-alpha1 PRKAA1 100 1000 JNK-IN-8 AMPK-alpha2 PRKAA2 84 1000 JNK-IN-8 ANKK1 ANKK1 75 1000 JNK-IN-8 ARK5 NUAK1 100 1000 JNK-IN-8 ASK1 MAP3K5 100 1000 JNK-IN-8 ASK2 MAP3K6 93 1000 JNK-IN-8 AURKA AURKA 100 1000 JNK-IN-8 AURKA AURKA 84 1000 JNK-IN-8 AURKB AURKB 83 1000 JNK-IN-8 AURKB AURKB 96 1000 JNK-IN-8 AURKC AURKC 95 1000 JNK-IN-8 -

Induction of the CD24 Surface Antigen in Primary Undifferentiated Human Adipose Progenitor Cells by the Hedgehog Signaling Pathway
Article Induction of the CD24 Surface Antigen in Primary Undifferentiated Human Adipose Progenitor Cells by the Hedgehog Signaling Pathway Francesco Muoio 1 , Stefano Panella 1 , Yves Harder 2,3 and Tiziano Tallone 1,* 1 Foundation for Cardiological Research and Education (FCRE), Cardiocentro Ticino, 6807 Taverne, Switzerland; [email protected] (F.M.); [email protected] (S.P.) 2 Department of Plastic, Reconstructive, and Aesthetic Surgery, EOC, 6900 Lugano, Switzerland; [email protected] or [email protected] 3 Faculty of Biomedical Sciences, Università della Svizzera Italiana, 6900 Lugano, Switzerland * Correspondence: [email protected] or [email protected]; Tel.: +41-91-805-3885 Abstract: In the murine model system of adipogenesis, the CD24 cell surface protein represents a valuable marker to label undifferentiated adipose progenitor cells. Indeed, when injected into the residual fat pads of lipodystrophic mice, these CD24 positive cells reconstitute a normal white adipose tissue (WAT) depot. Unluckily, similar studies in humans are rare and incomplete. This is because it is impossible to obtain large numbers of primary CD24 positive human adipose stem cells (hASCs). This study shows that primary hASCs start to express the glycosylphosphatidylinositol (GPI)-anchored CD24 protein when cultured with a chemically defined medium supplemented with molecules that activate the Hedgehog (Hh) signaling pathway. Therefore, this in vitro system may help understand the biology and role in adipogenesis of the CD24-positive hASCs. The induced Citation: Muoio, F.; Panella, S.; cells’ phenotype was studied by flow cytometry, Real-Time Quantitative Polymerase Chain Reaction Harder, Y.; Tallone, T. Induction of the (RT-qPCR) techniques, and their secretion profile. -

Human B Cells Marked by CD24 Expression Immunodeficiency
Characterization of Lymphocyte Subsets in Patients with Common Variable Immunodeficiency Reveals Subsets of Naive Human B Cells Marked by CD24 Expression This information is current as of September 29, 2021. Marcela Vlková, Eva Fronková, Veronika Kanderová, Ales Janda, Sárka Ruzicková, Jirí Litzman, Anna Sedivá and Tomás Kalina J Immunol 2010; 185:6431-6438; Prepublished online 1 November 2010; Downloaded from doi: 10.4049/jimmunol.0903876 http://www.jimmunol.org/content/185/11/6431 http://www.jimmunol.org/ Supplementary http://www.jimmunol.org/content/suppl/2010/11/01/jimmunol.090387 Material 6.DC1 References This article cites 34 articles, 15 of which you can access for free at: http://www.jimmunol.org/content/185/11/6431.full#ref-list-1 Why The JI? Submit online. by guest on September 29, 2021 • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology Characterization of Lymphocyte Subsets in Patients with Common Variable Immunodeficiency Reveals Subsets of Naive Human B Cells Marked by CD24 Expression Marcela Vlkova´,* Eva Fronˇkova´,†,‡ Veronika Kanderova´,†,‡ Alesˇ Janda,† Sˇa´rka Ru˚zˇicˇkova´,x Jirˇ´ı Litzman,* Anna Sˇediva´,{ and Toma´sˇ Kalina†,‡ Increased proportions of naive B cell subset and B cells defined as CD27negCD21negCD38neg are frequently found in patients with common variable immunodeficiency (CVID) syndrome. -

Relationship of CD44 CD24 Breast Cancer Stem Cells and Axillary Lymph Node Metastasis
Wei et al. Journal of Translational Medicine 2012, 10(Suppl 1):S6 http://www.translational-medicine.com/content/10/S1/S6 PROCEEDINGS Open Access Relationship of CD44+CD24-/low breast cancer stem cells and axillary lymph node metastasis Wei Wei1*, Hui Hu1, Haosheng Tan1, Louis WC Chow2, Adrian YS Yip2, Wings TY Loo2 From Organisation for Oncology and Translational Research (OOTR) 7th Annual Conference Hong Kong. 13-14 May 2011 Abstract Background: Axillary node staging plays an important role in the prognostic evaluation and planning of adjuvant treatment. Breast cancer stem cells, identified on the basis of CD44+CD24-/low expression, are associated with metastases and drug resistance. It is therefore important to investigate the proportion of CD44+CD24-/low breast cancer stem cells for the diagnosis of metastases in axillary nodes. Methods: Thirty-two ipsilateral axillary lymph nodes were collected from patients diagnosed with invasive breast cancer. Each lymph node (LN) was divided into two equals – one was examined by H&E staining, while the other was made into a single cell suspension to study the content of CD44+CD24-/low cells by flow cytometry (FCM). The relationship was investigated between the content of CD44+CD24-/low cells and metastases in axillary nodes which were confirmed by histology. Associations were tested using the chi-square test (linear-by-linear association), and the significance level was set at a value of p < 0.05. Results: In the 32 axillary nodes, the level of CD44+CD24-/low cells was determined to be between 0 and 18.4%: there was no presence of CD44+CD24-/low cells in 9 LNs, of which 2 had confirmed metastasis; there were less than 10% CD44+CD24-/low cells in 12 LNs, of which 6 had confirmed metastasis; and there were more than 10% CD44+CD24-/ low cells in 11 LNs, of which 9 had confirmed metastasis. -

CD24 Is a Novel Predictor for Poor Prognosis of Hepatocellular
Published OnlineFirst August 25, 2009; DOI: 10.1158/1078-0432.CCR-09-0151 Published Online First on August 25, 2009 as 10.1158/1078-0432.CCR-09-0151 Imaging, Diagnosis, Prognosis CD24 Is a Novel Predictor for Poor Prognosis of Hepatocellular Carcinoma after Surgery Xin-Rong Yang,1 Yang Xu,1 Bin Yu,2 Jian Zhou,1 Jia-Chu Li,3 Shuang-Jian Qiu,1 Ying-Hong Shi,1 Xiao-Ying Wang,1 Zhi Dai,1 Guo-Ming Shi,1 Bin Wu,1 Li-Ming Wu,1 Guo-Huan Yang,1 Bo-Heng Zhang,1 Wen-Xin Qin,2 and Jia Fan1,4 Abstract Purpose: To investigate the role of CD24 in tumor invasion and prognostic significance in hepatocellular carcinoma (HCC). Experimental Design: CD24 expression was measured in stepwise metastatic HCC cell lines, tumor, peritumoral tissues, and normal liver tissues by quantitative real-time PCR and Western blot. The role of CD24 in HCC was investigated by CD24 depletion using small interfering RNA. Tumor tissue microarrays of 314 HCC patients who underwent resection between 1997 and 2000 were used to detect expression of CD24, β-catenin, and proliferating cell nuclear antigen. Prognostic significance was assessed using Kaplan-Meier survival estimates and log-rank tests. Results: CD24 was overexpressed in the highly metastatic HCC cell line and in tumor tissues of patients with recurrent HCC. Depletion of CD24 caused a notable decrease in cell proliferation, migration, and invasiveness in vitro. Univariate and multivariate anal- yses revealed that CD24 was a significant predictor for overall survival and relapse-free survival. CD24 expression was correlated with poor prognosis independent of α-feto- protein, tumor-node-metastasis stage, and Edmondson stage. -

Understanding the Transport Mechanism of BBB Peptide Shuttles: Thrre and Miniap-4 As Case Studies
Understanding the transport mechanism of BBB peptide shuttles: THRre and MiniAp-4 as case studies Cristina Fuster Juncà ADVERTIMENT. La consulta d’aquesta tesi queda condicionada a l’acceptació de les següents condicions d'ús: La difusió d’aquesta tesi per mitjà del servei TDX (www.tdx.cat) i a través del Dipòsit Digital de la UB (diposit.ub.edu) ha estat autoritzada pels titulars dels drets de propietat intel·lectual únicament per a usos privats emmarcats en activitats d’investigació i docència. No s’autoritza la seva reproducció amb finalitats de lucre ni la seva difusió i posada a disposició des d’un lloc aliè al servei TDX ni al Dipòsit Digital de la UB. No s’autoritza la presentació del seu contingut en una finestra o marc aliè a TDX o al Dipòsit Digital de la UB (framing). Aquesta reserva de drets afecta tant al resum de presentació de la tesi com als seus continguts. En la utilització o cita de parts de la tesi és obligat indicar el nom de la persona autora. ADVERTENCIA. La consulta de esta tesis queda condicionada a la aceptación de las siguientes condiciones de uso: La difusión de esta tesis por medio del servicio TDR (www.tdx.cat) y a través del Repositorio Digital de la UB (diposit.ub.edu) ha sido autorizada por los titulares de los derechos de propiedad intelectual únicamente para usos privados enmarcados en actividades de investigación y docencia. No se autoriza su reproducción con finalidades de lucro ni su difusión y puesta a disposición desde un sitio ajeno al servicio TDR o al Repositorio Digital de la UB. -

Correlation of CD44 and CD24 Expression with the Positive Response After Neoadjuvant Chemotherapy in Stage IIIB Breast Cancer Pa
ORIGINAL ARTICLE Bali Medical Journal (Bali MedJ) 2021, Volume 10, Number 1: 99-102 P-ISSN.2089-1180, E-ISSN: 2302-2914 Correlation of CD44 and CD24 expression with the positive response after neoadjuvant Published by Bali Medical Journal chemotherapy in stage IIIB breast cancer patient at Dr. Saiful Anwar Hospital, Malang, Indonesia Muhammad Bachtiar Budianto1*, Artono Isharanto2, Andry Haris3 1Department of Oncology Surgery, ABSTRACT Faculty of Medicine, Universitas Brawijaya, Malang, Indonesia Background: Breast cancer is a type of cancer due to abnormal cell growth of breast tissue. CD44 and CD24 is a potential 2 Department of Thoracic and target in breast cancer therapy. This study aims to evaluate the responsiveness of stage 3 breast cancer to chemotherapy by Cardiovascular Surgery, Faculty of measuring the expression levels of CD44 and CD24 molecules. Medicine, Universitas Brawijaya, Malang, Indonesia Methods: We conducted an observational study with pre-and –post-intervention or test research types. This study was 3General Surgery Resident, Faculty of performed on 49 Luminal Stadium IIIB subtype breast cancer patients who received 3 series of neoadjuvant chemotherapy at Medicine, Universitas Brawijaya, Malang, RSUD dr. Saiful Anwar Malang, immunohistochemical examination, and painting of tissue specimens from the biopsy results. Indonesia Data were analyzed using SPSS version 20 for Windows. Results: Based on the Paired t-test and discriminant analysis, there were no significant differences between CD44 and CD24 expression before and after chemotherapy (p=0.501 and p=0.097, respectively). Therefore, the results of the study prompt *Corresponding author: that the expression of CD44 and CD24 could not be a predictor of the chemotherapy response.