(Rodentia, Sigmodontinae) from Southern Brazil Atlantic Forest
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Ectoparasites of Wild Rodents from Parque Estadual Da Cantareira (Pedra Grande Nuclei), São Paulo, Brazil
ECTOPARASITES OF WILD RODENTS FROM PARQUE ESTADUAL DA CANTAREIRA (PEDRA GRANDE NUCLEI), SÃO PAULO, BRAZIL FERNANDA A. NIERI-BASTOS1 DARCI M. BARROS-BATTESTI1 PEDRO M. LINARDI2 MARCOS AMAKU3 ARLEI MARCILI4 SANDRA E. FAVORITO5; RICARDO PINTO-DA-ROCHA6 ABSTRACT:- NERI-BASTOS, F.A.; BARROS-BATTESTI, D.M.; LINARDI, P.M.; AMAKU, M.; MARCILI, A.; FAVORITO, S.E.; PINTO-DA-ROCHA, R. Ectoparasites of wild rodents from Parque Estadual da Cantareira (Pedra Grande Nuclei), São Paulo, Brazil. [Ectoparasitos de roedores silvestres do Parque Estadual da Cantareira (Núcleo Pedra Grande), São Paulo, Brasil.] Revista Brasileira de Parasitologia Veterinária, v. 13, n. 1, p. 29-35, 2004. Laboratório de Parasitologia, Instituto Butantan, Av. Vital Brasil 1500, São Paulo, SP 05503-900, Brazil. E-mail: [email protected] Sixteen ectoparasite species were collected from 195 wild rodents, between February 2000 and January 2001, in an Ecological Reserve area of the Parque Estadual da Cantareira, in the municipalities of Caieiras, Mariporã and Guarulhos, State of São Paulo, Brazil. Fifty three percent of the captured rodents were found infested, with the highest prevalences observed for the mites Gigantolaelaps gilmorei and G. oudemansi on Oryzomys russatus; G. wolffsohni, Lalelaps paulistanensis and Mysolaelaps parvispinosus on Oligoryzomys sp. In relation to the fleas, Polygenis (Neopolygenis) atopus presented the highest prevalence, infesting Oryzomys russatus. The highest specificity indices were found for Eubrachylaelaps rotundus/Akodon sp.; G. gilmorei and G. oudemansi/O. russatus; and Laelaps navasi/Juliomys pictipes. When average infestation intensities were related to specificity indices, the relationship was only significant for Brucepattersonius sp. and O. russatus (P<0.05). -

Sobre a BIOTA NEOTROPICA
Biota Neotrop. vol. 9, no. 1, jan./mar. 2009 ISSN 1678-6424 ISSN 1678-6424 vol. 9, no. 1, jan./mar. 2009 jan./mar. 1, no. 9, vol. Sumário Artigos Uma revisão da distribuição de Ocotea curucutuensis J.B. Baitello na região sudeste do Brasil vol. 9, no. 1, jan./mar. 2009 Frederico Alexandre Roccia Dal Pozzo Arzolla, João Batista Baitello, George John Shepherd, Gláucia Cortez Ramos de Paula & Ricardo Bertoncello ...............................................21 Novos registros, sinonímia e descrição do macho de Culicoides horticola Lutz (Diptera: Ceratopogonidae) Maria Luiza Felippe-Bauer & Gustavo Ricardo Spinelli .....................................................................................................................................................................................................27 Os pequenos mamíferos da altamente impactada Floresta Atlântica do Nordeste do Brasil, Centro de Endemismo Pernambuco Paulo Henrique Asfora & Antonio Rossano Mendes Pontes ..............................................................................................................................................................................................31 Ácaros associados ao cafeeiro (Coffea spp.) no estado de São Paulo, Brasil. Parte I. Mesostigmata Jeferson Luiz de Carvalho Mineiro, Adalton Raga, Mario Eidi Sato & Antonio Carlos Lofego ...........................................................................................................................................37 Relações entre diversidade íctia e fatores hidrodinâmicos -

Rodentia: Cricetidae: Sigmodontinae) in São Paulo State, Southeastern Brazil: a Locally Extinct Species?
Volume 55(4):69‑80, 2015 THE PRESENCE OF WILFREDOMYS OENAX (RODENTIA: CRICETIDAE: SIGMODONTINAE) IN SÃO PAULO STATE, SOUTHEASTERN BRAZIL: A LOCALLY EXTINCT SPECIES? MARCUS VINÍCIUS BRANDÃO¹ ABSTRACT The Rufous-nosed Mouse Wilfredomys oenax is a rare Sigmodontinae rodent known from scarce records from northern Uruguay and south and southeastern Brazil. This species is under- represented in scientific collections and is currently classified as threathened, being considered extinct at Curitiba, Paraná, the only confirmed locality of the species at southeastern Brazil. Although specimens from São Paulo were already reported, the presence of this species in this state seems to have passed unnoticed in recent literature. Through detailed morphological ana- lyzes of specimens cited in literature, the present work confirms and discusses the presence of this species in São Paulo state from a specimen collected more than 70 years ago. Recently, by the use of modern sampling methods, other rare Sigmodontinae rodents, such as Abrawayomys ruschii, Phaenomys ferrugineous and Rhagomys rufescens, have been recorded to São Paulo state. However, no specimen of Wilfredomys oenax has been recently reported indicating that this species might be locally extinct. The record mentioned here adds another species to the state of São Paulo mammal diversity and reinforces the urgency of studying Wilfredomys oenax. Key-Words: Atlantic Forest; Scientific collection; Threatened species. INTRODUCTION São Paulo is one the most studied states in Brazil regarding to fauna. Mammal lists from this state have Mammal species lists based on voucher-speci- been elaborated since the late XIX century (Von Iher- mens and literature records are essential for offering ing, 1894; Vieira, 1944a, b, 1946, 1950, 1953; Vivo, groundwork to understand a species distribution and 1998). -

With Focus on the Genus Handleyomys and Related Taxa
Brigham Young University BYU ScholarsArchive Theses and Dissertations 2015-04-01 Evolution and Biogeography of Mesoamerican Small Mammals: With Focus on the Genus Handleyomys and Related Taxa Ana Villalba Almendra Brigham Young University - Provo Follow this and additional works at: https://scholarsarchive.byu.edu/etd Part of the Biology Commons BYU ScholarsArchive Citation Villalba Almendra, Ana, "Evolution and Biogeography of Mesoamerican Small Mammals: With Focus on the Genus Handleyomys and Related Taxa" (2015). Theses and Dissertations. 5812. https://scholarsarchive.byu.edu/etd/5812 This Dissertation is brought to you for free and open access by BYU ScholarsArchive. It has been accepted for inclusion in Theses and Dissertations by an authorized administrator of BYU ScholarsArchive. For more information, please contact [email protected], [email protected]. Evolution and Biogeography of Mesoamerican Small Mammals: Focus on the Genus Handleyomys and Related Taxa Ana Laura Villalba Almendra A dissertation submitted to the faculty of Brigham Young University in partial fulfillment of the requirements for the degree of Doctor of Philosophy Duke S. Rogers, Chair Byron J. Adams Jerald B. Johnson Leigh A. Johnson Eric A. Rickart Department of Biology Brigham Young University March 2015 Copyright © 2015 Ana Laura Villalba Almendra All Rights Reserved ABSTRACT Evolution and Biogeography of Mesoamerican Small Mammals: Focus on the Genus Handleyomys and Related Taxa Ana Laura Villalba Almendra Department of Biology, BYU Doctor of Philosophy Mesoamerica is considered a biodiversity hot spot with levels of endemism and species diversity likely underestimated. For mammals, the patterns of diversification of Mesoamerican taxa still are controversial. Reasons for this include the region’s complex geologic history, and the relatively recent timing of such geological events. -

Pontificia Universidad Católica Del Ecuador
CORE Metadata, citation and similar papers at core.ac.uk Provided by Repositorio Digital PUCE PONTIFICIA UNIVERSIDAD CATÓLICA DEL ECUADOR FACULTAD DE CIENCIAS EXACTAS Y NATURALES ESCUELA DE BIOLOGÍA Genetic and morphological variability of the páramo Oldfield mouse Thomasomys paramorum Thomas, 1898 (Rodentia: Cricetidae): evidence for a complex of species Tesis previa a la obtención del título de Magister en Biología de la Conservación CARLOS ESTEBAN BOADA TERÁN Quito, 2013 II Certifico que la Tesis de Maestría en Biología de la Conservación del candidato Carlos Esteban Boada Terán ha sido concluida de conformidad con las normas establecidas; por tanto, puede ser presentada para la calificación correspondiente. Dr. Omar Lenin Torres Carvajal Director de Tesis Mayo de 2013 III Dedicado a mi hijo Joaquín IV ACKNOWLEDGMENTS This manuscript was presented as a requirement for graduation at Pontificia Universidad Católica del Ecuador, Master´s program in Conservation Biology. I thank O. Torres- Carvajal for his mentorship, J. Patton for reviewing the first draft of the manuscript and provide valuable suggestions and comments, and S. Burneo for allowing the examination of specimens deposited at Museo de Zoología (QCAZ), sección Mastozoología. For field support I thank Viviana Narváez, Daniel Chávez, Roberto Carrillo, Simón Lobos, Julia Salvador, Adriana Argoti and Amy Scott. Finally I am grateful to Mary Eugenia Ordóñez, Gaby Nichols, Andrea Manzano and Diana Flores for their help in the laboratory. I thank to SENESCYT because most laboratory equipment was purchased with the project "Inventory and Morphological Characterization and Genetic Diversity of Amphibians, Reptiles and Birds of the Andes of Ecuador", code PIC-08-0000470. -

Proquest Dissertations
The Neotropical rodent genus Rhipidom ys (Cricetidae: Sigmodontinae) - a taxonomic revision Christopher James Tribe Thesis submitted for the degree of Doctor of Philosophy University College London 1996 ProQuest Number: 10106759 All rights reserved INFORMATION TO ALL USERS The quality of this reproduction is dependent upon the quality of the copy submitted. In the unlikely event that the author did not send a complete manuscript and there are missing pages, these will be noted. Also, if material had to be removed, a note will indicate the deletion. uest. ProQuest 10106759 Published by ProQuest LLC(2016). Copyright of the Dissertation is held by the Author. All rights reserved. This work is protected against unauthorized copying under Title 17, United States Code. Microform Edition © ProQuest LLC. ProQuest LLC 789 East Eisenhower Parkway P.O. Box 1346 Ann Arbor, Ml 48106-1346 ABSTRACT South American climbing mice and rats, Rhipidomys, occur in forests, plantations and rural dwellings throughout tropical South America. The genus belongs to the thomasomyine group, an informal assemblage of plesiomorphous Sigmodontinae. Over 1700 museum specimens were examined, with the aim of providing a coherent taxonomic framework for future work. A shortage of discrete and consistent characters prevented the use of strict cladistic methodology; instead, morphological assessments were supported by multivariate (especially principal components) analyses. The morphometric data were first assessed for measurement error, ontogenetic variation and sexual dimorphism; measurements with most variation from these sources were excluded from subsequent analyses. The genus is characterized by a combination of reddish-brown colour, long tufted tail, broad feet with long toes, long vibrissae and large eyes; the skull has a small zygomatic notch, squared or ridged supraorbital edges, large oval braincase and short palate. -

Chec List ISSN 1809-127X (Available at Journal of Species Lists and Distribution
Check List 8(3): 557-559, 2012 © 2012 Check List and Authors Chec List ISSN 1809-127X (available at www.checklist.org.br) Journal of species lists and distribution N Rhagomys rufescens Sigmodontinae) Thomas, 1886 ISTRIBUTIO New distribution reports of (Rodentia: D * and Maria Rita Silvério Pires RAPHIC G Caryne Aparecida de Carvalho Braga EO G N Universidade Federal de Ouro Preto, Departamento de Evolução, Biodiversidade e Meio Ambiente, Laboratório de Zoologia dos Vertebrados, O [email protected] Campus Morro do Cruzeiro. CEP 35400-000. Ouro Preto, MG, Brasil. * Corresponding author. E-mail: OTES N Abstract: Rhagomys rufescens is a rare, arboreal sigmodontine rodent endemic to the Brazilian Atlantic Forest biome. This species is known from eight localities in Brazil. Here we present a new report based on four individuals of this species registered in Serra do Ouro Branco, municipality of Ouro Branco (Minas Gerais, Brazil). One juvenile male, one adult male ofand the two last juvenile registered females location. were captured in pitfall traps during the rainy season, in a study of small mammal ecology. This is the first record for the Espinhaço Mountain range and the northernmost report for the species in this state, 85 km northeast Rhagomys rufescens (Thomas, 1886) is an enigmatic level as “incertae sedis” (Reig 1980; 1984; McKenna and Forest domain, at the south of the Espinhaço mountain arboreal sigmodontine rodent classified at the suprageneric range. Our report occurred in an area of vegetation characterized as semi-deciduous mountain forest. A Bell 1997; Smith and Patton 1999; Musser and Carleton portion of the sampled locality is now inside a conservation 2005). -

Ecog-03504.Pdf
Ecography ECOG-03504 Bovendorp, R. S., Brum, F. T., McCleery, R. A., Baiser, B., Loyola, R., Cianciaruso, M. V. and Galetti, M. 2018. Defaunation and fragmentation erode small mammal diversity dimensions in tropical forests. – Ecography doi: 10.1111/ecog.03504 Supplementary material Supplemental material Appendix 1: Studies included in the index of defaunation analyses. REFERENCES Agência Estadual de Meio Ambiente e Recursos Hídricos 2006. Plano de Manejo Estação Ecológica de Caetés. — CPRH. Albuquerque, H. G. et al. 2013. Mammals of a forest fragment in Cambuci municipality, state of Rio de Janeiro, Brazil. — Check List 9: 1505-1509. Antunes, A. Z. and Eston, M. R. 2009. Mamíferos (chordata: mammalia) florestais de médio e grande porte registrados em Barreiro Rico, Anhembi, Estado de São Paulo —Revista do Instituto Florestal 21: 201-215. Aximoff, I. et al. 2015. Long-term survey by camera traps of non-volant mammals in two national parks in Rio de Janeiro State. — Oecologia Australis 19: 215-231. Bariani, M. D. 2010. Impacto da colheita de cana-de-açúcar nos mamíferos e répteis silvestres da região do Pontal do Paranapanema, SP. Instituto de Pesquisas Ecológicas - IPE, p 50. Bastiani, E. 2015. Levantamento preliminar de mamíferos de médio e grande porte na Floresta Nacional de Piraí do Sul, Paraná, Brasil. Conference poster. XI Congresso de Ecologia do Brasil. Bastiani, E. et al. 2015. Felinos da Floresta Nacional de Piraí do Sul, Paraná-Brasil. — Acta Zoológica Mexicana 31: 23-26. Bernardo, C. S. S. 2004. Abundância, densidade e tamanho populacional de aves e mamíferos cinegéticos no Parque Estadual Ilha do Cardoso, SP, Brasil. -

DE Thomasomys Y DESCRIPCIÓN DE UNA NUEVA ESPECIE (RODENTIA, CRICETIDAE)
Mastozoología Neotropical, 26(2):308-330 Mendoza, 2019 Copyright © SAREM, 2019 Versión on-line ISSN 1666-0536 hp://www.sarem.org.ar hps://doi.org/10.31687/saremMN.19.26.2.0.04 hp://www.sbmz.org Artículo DIVERSIDAD INSOSPECHADA EN LOS ANDES DE ECUADOR: FILOGENIA DEL GRUPO “cinereus” DE Thomasomys Y DESCRIPCIÓN DE UNA NUEVA ESPECIE (RODENTIA, CRICETIDAE) Jorge Brito1, Nicolás Tinoco2, Jenny Curay1, Rocío Vargas1, Carolina Reyes-Puig3, Víctor Romero4,5 y Ulyses F. J. Pardiñas1, 6 1 Instituto Nacional de Biodiversidad (INABIO), Quito, Ecuador. [Correspondencia: Jorge Brito <[email protected]>] 2 Museo de Zoología, Escuela de Biología, Ponticia Universidad Católica del Ecuador, Quito, Ecuador. 3 Instituto de Zoología Terrestre, Museo de Zoología, Instituto BIOSFERA, Colegio de Ciencias Biológicas y Ambientales, Universidad San Francisco de Quito (USFQ) Cumbayá, Quito, Ecuador. 4 Museo de Zoología, Universidad Técnica Particular de Loja, San Cayetano Alto, Loja, Ecuador. 5 Postgrado en Ciencias Biológicas, Departamento de Estudios Ambientales, Universidad Simón Bolívar, Caracas, Venezuela. 6 Instituto de Diversidad y Evolución Austral (IDEAus – CONICET), Puerto Madryn, Chubut, Argentina. RESUMEN. Con base en ejemplares recolectados en los Andes surorientales de Ecuador, Parque Nacional Sangay, pero también integrando otros materiales de colecciones, se efectuó una revisión del grupo “cinereus” del género de cricétidos Thomasomys Coues, 1884 (Sigmodontinae, Thomasomyini). Como resultado de dicha revisión, además de proponerse una hipótesis -
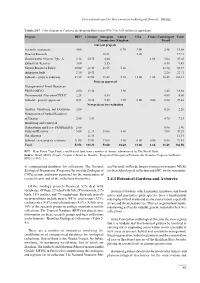
CBD First National Report
First national report for the Convention on Biological Diversity - BRAZIL Tabela 2-37. Pilot Program to Conserve the Brazilian Rain Forest PPG-7 (In US$ million or equivalent). Projects RFT* Germany European United USA France Counterpart Total Commission Kingdom Brazil Current projects Scientific institutions 9.00 0.70 3.00 2.98 15.68 Directed Research 10.91 9.00 19.91 Demonstrative Projects Type A 3.18 20.75 4.44 1.68 3.00 33.05 Extractivist Reserves 3.00 5.55 0.90 9.45 Natural Resources Policy 20.00 28.48 18.55 5.00 11.40 83.43 Indigenous lands 2.10 18.41 2.20 22.71 Subtotal – projects underway 37.28 67.64 39.45 5.70 12.00 1.68 20.48 184.23 Projects approved Management of Forest Resources- PROMANEJO 2.00 13.54 1.90 1.40 18.84 Environmental Education-CEDUC 2.25 5.55 0.80 8.60 Subtotal - projects approved 4.25 13.54 5.55 1.90 0.00 0.00 2.20 27.44 New projects for evaluation Analysis, Monitoring and Evaluation 2.00 0.20 2.20 Management of Natural Resources of Várzeas 2.00 4.54 0.70 7.24 Monitoring and Control of Deforestation and Fires- PRODESQUE 2.00 0.90 2.90 Parks and Reserves 5.00 21.15 13.00 3.00 7.00 49.15 Not allocated 11.34 11.34 Subtotal - new projects (estimate) 11.00 37.03 13.00 3.00 0.00 0.00 8.80 72.83 Total 52.53 118.21 58.00 10.60 12.00 1.68 31.48 284.50 RFT = Rain Forest Trust Fund, a multilateral fund from a number of donors, administered by The World Bank. -

Trypanosoma Cruzi Transmission in the Wild and Its Most Important
Jansen et al. Parasites & Vectors (2018) 11:502 https://doi.org/10.1186/s13071-018-3067-2 REVIEW Open Access Trypanosoma cruzi transmission in the wild and its most important reservoir hosts in Brazil Ana Maria Jansen*, Samanta Cristina das Chagas Xavier and André Luiz Rodrigues Roque Abstract Trypanosoma cruzi (Kinetoplastea: Trypanosomatidae) infects all tissues of its hosts, which along with humans, include hundreds of mammalian species in the Americas. The epidemiology of T. cruzi has been changing in that currently the majority of the cases and/or outbreaks of Chagas disease occur by the ingestion of comestibles contaminated by T. cruzi metacyclic forms. These cases/outbreaks occur in distinct regional scenarios, mainly in the Amazon biome and are related to the local interaction mode of humans with their surroundings, as well as with the overall local ecological peculiarities. As trypanosomiasis caused by T. cruzi is primarily a zoonosis, understanding the variables that influences its transmission in the wild as well as the role played by the extant fauna in the maintenance of the parasite, is critical in establishing control measures. Here, we present the results of our studies of T. cruzi infection of free ranging wild mammalian fauna in the five biomes of Brazil, a country of continental dimensions. From 1992 up to 2017, we examined a total of 6587 free-ranging non-volant wild mammal specimens. Our studies found that 17% of mammals were seropositive and 8% of all animals displayed positive hemocultures indicative of high parasitemia and, consequently, of infectivity potential. We observed that opossums, mainly Philander spp. -

Wiedomys Cerradensis Gonçalves, Almeida and Bonvicino, 2005 (Mammalia: Rodentia: Cricetidae) in Tocantins And
Check List 9(3): 680–683, 2013 © 2013 Check List and Authors Chec List ISSN 1809-127X (available at www.checklist.org.br) Journal of species lists and distribution N Wiedomys cerradensis Gonçalves, Almeida and Bonvicino, 2005 (Mammalia: Rodentia: Cricetidae) in Tocantins and ISTRIBUTIO Goiás states, central-northern Brazil D 1* 2 2,3 1 RAPHIC Alexandra M. R. Bezerra , Ana Lazar , Cibele R. Bonvicino and Jader Marinho-Filho G EO G 1 Universidade de Brasília ‘Campus Dacy Ribeiro’, Instituto de Biologia, Departamento de Zoologia, Coleção de Mamíferos. Asa Norte. CEP 70910- N O 900. Brasília, DF, Brazil. 2 Instituto Oswaldo Cruz, Fiocruz, Laboratório de Biologia e Parasitologia de Mamíferos Silvestres Reservatórios. Avenida Brasil, 4365. CEP 21045- 900. Rio de Janeiro, RJ, Brazil. OTES 3 Instituto Nacional de Câncer. Programa de Genética, CPQ. Rio de Janeiro, RJ, Brazil. N * Corresponding author. E-mail: [email protected] Abstract: We report two new localities for the rodent species Wiedomys cerradensis Gonçalves, Almeida and Bonvicino, 2005, previously known only from its type locality, in southwestern Bahia state, and one neighbor locality. Three new specimens were collected along the Paranã River Valley, in the Goiás and Tocantins states, central Brazil. These records extend the range of this species to the northwest, approximately 190 km. We provide external and cranial measurements of these specimens and comment on their morphology. Wiedomys Hershkovitz, 1959 is a rodent genus endemic Wiedomys cerradensis of Brazilian semiarid with two recognized species: W. the UICN (Bonvicino and Marinho-Filho 2008) due to pyrrhorhinus (Wied-Neuwied, 1821) and W. cerradensis its recent description and is limitedlisted as information Data Deficient on itsby Gonçalves, Almeida and Bonvicino, 2005.