Using Deeplabcut for 3D Markerless Pose Estimation Across Species and Behaviors
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Interpretable and Comparable Measurement for Computational Ethology
Interpretable and Comparable Measurement for Computational Ethology The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Urban, Konrad N. 2020. Interpretable and Comparable Measurement for Computational Ethology. Bachelor's thesis, Harvard College. Citable link https://nrs.harvard.edu/URN-3:HUL.INSTREPOS:37364750 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA Interpretable and Comparable Measurement for Computational Ethology By Konrad Urban Adviser: Venkatesh N. Murthy Thesis Advisor: Venkatesh N. Murthy Konrad Urban Interpretable and Comparable Measurement for Computational Ethology Abstract Computational ethology, the field of automated behavioral analysis, has introduced a number of new techniques in recent years, including supervised, unsupervised, and statistical approaches for measuring behavior. These approaches have made the measurement of behavior more precise and efficient, improving significantly on human annotation of behavior. Despite this improvement, however, researchers in the field often interpret and report results primarily with reference to traditional, linguistic categories of behavior, which reduces measurement precision. I argue that using interpretable measurement techniques can help researchers to overcome this imprecision. Further, I present Basin, a system for performing interpretable behavioral measurement. Pioneering work has recently developed deep learning systems which can accurately estimate the pose of animals in videos. Basin builds on this work by creating a system in which researchers can construct behavioral measurements which are functions of an animal's pose. -

Supplementary Material for “Pretraining Boosts Out-Of-Domain Robustness for Pose Estimation”
Supplementary Material for “Pretraining boosts out-of-domain robustness for pose estimation” Alexander Mathis1;2∗y Thomas Biasi2∗ Steffen Schneider1;3 Mert Yuksekg¨ on¨ ul¨ 2;3 Byron Rogers4 Matthias Bethge3 Mackenzie W. Mathis1;2z 1EPFL 2Harvard University 3University of Tubingen¨ 4Performance Genetics ∗Equal contribution. yCorrespondence: [email protected] zCorrespondence: [email protected] 1. Additional information on the Horse-10 dataset The table lists the following statistics: labeled frames, scale (nose-to-eye distance in pixels), and whether a horse was within domain (w.d) or out-of-domain (o.o.d.) for each shuffle. Horse Identifier samples nose-eye dist shuffle 1 shuffle 2 shuffle 3 BrownHorseinShadow 308 22.3 o.o.d o.o.d w.d. BrownHorseintoshadow 289 17.4 o.o.d o.o.d o.o.d Brownhorselight 306 15.57 o.o.d w.d. o.o.d Brownhorseoutofshadow 341 16.22 o.o.d w.d. w.d. ChestnutHorseLight 318 35.55 w.d. w.d. o.o.d Chestnuthorseongrass 376 12.9 o.o.d w.d. w.d. GreyHorseLightandShadow 356 14.41 w.d. w.d. o.o.d GreyHorseNoShadowBadLight 286 16.46 w.d. o.o.d w.d. TwoHorsesinvideobothmoving 181 13.84 o.o.d o.o.d w.d. Twohorsesinvideoonemoving 252 16.51 w.d. w.d. w.d. Sample1 174 24.78 o.o.d o.o.d o.o.d Sample2 330 16.5 o.o.d o.o.d o.o.d Sample3 342 16.08 o.o.d o.o.d o.o.d Sample4 305 18.51 o.o.d o.o.d w.d. -

Deep Learning Tools for the Measurement of Animal Behavior In
Deep learning tools for the measurement of animal behavior in neuroscience Mackenzie Weygandt Mathis1,* and Alexander Mathis1 1Harvard University, Cambridge, MA USA | *[email protected] Recent advances in computer vision have made accurate, fast behavior over long time periods into meaningful met- and robust measurement of animal behavior a reality. In the rics? How does one use behavioral quantification to build a past years powerful tools specifically designed to aid the mea- better understanding of the brain and an animal’s umwelt (1)? surement of behavior have come to fruition. Here we discuss how capturing the postures of animals - pose estimation - has In this review we discuss the advances, and challenges, in an- been rapidly advancing with new deep learning methods. While imal pose estimation and its impact on neuroscience. Pose es- challenges still remain, we envision that the fast-paced develop- timation refers to methods for measuring posture, while pos- ment of new deep learning tools will rapidly change the land- scape of realizable real-world neuroscience. ture denotes to the geometrical configuration of body parts. While there are many ways to record behavior (7–9), videog- Highlights: raphy is a non-invasive way to observe the posture of ani- mals. Estimated poses across time can then, depending on 1. Deep neural networks are shattering performance the application, be transformed into kinematics, dynamics, benchmarks in computer vision for various tasks. and actions (3,5–7, 10). Due to the low-dimensional nature of posture, these applications are computationally tractable. 2. Using modern deep learning approaches (DNNs) in the lab is a fruitful approach for robust, fast, and efficient A very brief history of pose estimation. -

Real-Time, Low-Latency Closed-Loop Feedback Using Markerless Posture
TOOLS AND RESOURCES Real-time, low-latency closed-loop feedback using markerless posture tracking Gary A Kane1, Gonc¸alo Lopes2, Jonny L Saunders3, Alexander Mathis1,4, Mackenzie W Mathis1,4* 1The Rowland Institute at Harvard, Harvard University, Cambridge, United States; 2NeuroGEARS Ltd, London, United Kingdom; 3Institute of Neuroscience, Department of Psychology, University of Oregon, Eugene, United States; 4Center for Neuroprosthetics, Center for Intelligent Systems, & Brain Mind Institute, School of Life Sciences, Swiss Federal Institute of Technology (EPFL), Lausanne, Switzerland Abstract The ability to control a behavioral task or stimulate neural activity based on animal behavior in real-time is an important tool for experimental neuroscientists. Ideally, such tools are noninvasive, low-latency, and provide interfaces to trigger external hardware based on posture. Recent advances in pose estimation with deep learning allows researchers to train deep neural networks to accurately quantify a wide variety of animal behaviors. Here, we provide a new DeepLabCut-Live! package that achieves low-latency real-time pose estimation (within 15 ms, >100 FPS), with an additional forward-prediction module that achieves zero-latency feedback, and a dynamic-cropping mode that allows for higher inference speeds. We also provide three options for using this tool with ease: (1) a stand-alone GUI (called DLC-Live!XGUI), and integration into (2) Bonsai, and (3) AutoPilot. Lastly, we benchmarked performance on a wide range of systems so that experimentalists can easily decide what hardware is required for their needs. *For correspondence: [email protected] Introduction In recent years, advances in deep learning have fueled sophisticated behavioral analysis tools Competing interest: See (Insafutdinov et al., 2016; Newell et al., 2016; Cao et al., 2017; Mathis et al., 2018b; page 26 Pereira et al., 2019; Graving et al., 2019; Zuffi et al., 2019). -

Sciencedirect.Com Current Opinion in Sciencedirect Neurobiology
Available online at www.sciencedirect.com Current Opinion in ScienceDirect Neurobiology Measuring and modeling the motor system with machine learning Sebastien B. Hausmanna, Alessandro Marin Vargasa, Alexander Mathisb and Mackenzie W. Mathisb Abstract constitute actions, motion, and what we perceive as the The utility of machine learning in understanding the motor primary output of the motor system. Now with deep system is promising a revolution in how to collect, measure, learning, the ability to measure these postures has and analyze data. The field of movement science already been massively facilitated. But the measurement of elegantly incorporates theory and engineering principles to movement is not the sole realm where machine learning guide experimental work, and in this review we discuss the has increased our ability to understand the motor growing use of machine learning: from pose estimation, kine- system. Modeling the motor system is also leveraging matic analyses, dimensionality reduction, and closed-loop new machine learning tools. This opens up new avenues feedback, to its use in understanding neural correlates and where neural, behavioral, and muscle activity data can untangling sensorimotor systems. We also give our perspec- be measured, manipulated, and modeled with ever- tive on new avenues, where markerless motion capture com- increasing precision and scale. bined with biomechanical modeling and neural networks could be a new platform for hypothesis-driven research. Measuring motor behavior Motor behavior remains the ultimate readout of internal Addresses EPFL, Swiss Federal Institute of Technology, Lausanne, Switzerland intentions. It emerges from a complex hierarchy of motor control circuits in both the brain and spinal cord, with Corresponding authors: Alexander Mathis (alexander.mathis@epfl. -

Measuring and Modeling the Motor System with Machine Learning
Measuring and modeling the motor system with machine learning Sébastien B. Hausmann1,†, Alessandro Marin Vargas1,†, Alexander Mathis1,*,& Mackenzie W. Mathis1,* 1EPFL, Swiss Federal Institute of Technology, Lausanne, Switzerland | †co-first *co-senior The utility of machine learning in understanding the motor sys- control circuits in both the brain and spinal cord, with the tem is promising a revolution in how to collect, measure, and ultimate output being muscles. Where these motor com- analyze data. The field of movement science already elegantly mands arise and how they are shaped and adapted over incorporates theory and engineering principles to guide exper- time still remains poorly understood. For instance, locomo- imental work, and in this review we discuss the growing use tion is produced by central pattern generators in the spinal of machine learning: from pose estimation, kinematic analy- cord and is influenced by goal-directed descending com- ses, dimensionality reduction, and closed-loop feedback, to its mand neurons from the brainstem and higher motor areas use in understanding neural correlates and untangling sensori- motor systems. We also give our perspective on new avenues to adopt specific gaits and speeds—the more complex and where markerless motion capture combined with biomechani- goal-directed the movement, the larger number of neural cir- cal modeling and neural networks could be a new platform for cuits recruited (5,6). Of course, tremendous advances have hypothesis-driven research. already been made in untangling the functions of motor cir- cuits since the times of Sherrington (7), but new advances in Highlights: machine learning for modeling and measuring the motor sys- tem will allow for better automation and precision, and new 1. -

Multi-Animal Pose Estimation and Tracking with Deeplabcut
bioRxiv preprint doi: https://doi.org/10.1101/2021.04.30.442096; this version posted April 30, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. Multi-animal pose estimation and tracking with DeepLabCut Jessy Lauer1,2, Mu Zhou1, Shaokai Ye1, William Menegas3, Tanmay Nath2, Mohammed Mostafizur Rahman4, Valentina Di Santo5,6, Daniel Soberanes2, Guoping Feng3, Venkatesh N. Murthy4, George Lauder5, Catherine Dulac4, Mackenzie W. Mathis 1,2 †, and Alexander Mathis 1,2,4 † 1Center for Neuroprosthetics, Center for Intelligent Systems, & Brain Mind Institute, School of Life Sciences, Swiss Federal Institute of Technology (EPFL), Lausanne, Switzerland 2Rowland Institute, Harvard University, Cambridge, MA USA 3Department of Brain and Cognitive Sciences and McGovern Institute for Brain Research, Massachusetts Institute of Technology, Cambridge, MA USA 4Department for Molecular Biology and Center for Brain Science, Harvard University, Cambridge, MA USA 5Department of Organismic and Evolutionary Biology, Harvard University, Cambridge, MA USA 6Department of Zoology, Stockholm University, Stockholm, Sweden †co-directed the work, mackenzie.mathis@epfl.ch, alexander.mathis@epfl.ch Estimating the pose of multiple animals is a challenging com- lar individuals. Here, many solutions have been proposed, puter vision problem: frequent interactions cause occlusions such as part affinity fields (7), associative embeddings (8, 9), and complicate the association of detected keypoints to the cor- transformers (10) and other mechanisms (11, 12). These are rect individuals, as well as having extremely similar looking ani- called bottom-up approaches, as detections and links are pre- mals that interact more closely than in typical multi-human sce- dicted from the image and the individuals are then “assem- narios. -

Dimensional Symmetries Underlying Mammalian Grid-Cell Activity Patterns
Probable nature of higher- dimensional symmetries underlying mammalian grid-cell activity patterns The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Mathis, Alexander, Martin B Stemmler, and Andreas VM Herz. 2015. “Probable nature of higher-dimensional symmetries underlying mammalian grid-cell activity patterns.” eLife 4 (1): e05979. doi:10.7554/eLife.05979. http://dx.doi.org/10.7554/eLife.05979. Published Version doi:10.7554/eLife.05979 Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:17295604 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA RESEARCH ARTICLE elifesciences.org Probable nature of higher-dimensional symmetries underlying mammalian grid- cell activity patterns Alexander Mathis1,2*, Martin B Stemmler3,4, Andreas VM Herz3,4 1Department of Molecular and Cellular Biology, Harvard University, Cambridge, United States; 2Center for Brain Science, Harvard University, Cambridge, United States; 3Bernstein Center for Computational Neuroscience, Munich, Germany; 4Fakultat¨ fur¨ Biologie, Ludwig-Maximilians-Universitat¨ Munchen,¨ Munich, Germany Abstract Lattices abound in nature—from the crystal structure of minerals to the honey-comb organization of ommatidia in the compound eye of insects. These arrangements provide solutions for optimal packings, efficient resource distribution, and cryptographic protocols. Do lattices also play a role in how the brain represents information? We focus on higher-dimensional stimulus domains, with particular emphasis on neural representations of physical space, and derive which neuronal lattice codes maximize spatial resolution. -

Pretraining Boosts Out-Of-Domain Robustness for Pose Estimation
Pretraining boosts out-of-domain robustness for pose estimation Alexander Mathis1;2*† Thomas Biasi2* Steffen Schneider1;3 Mert Yüksekgönül2;3 Byron Rogers4 Matthias Bethge3 Mackenzie W. Mathis1;2‡ 1EPFL 2Harvard University 3University of Tübingen 4Performance Genetics Abstract Transfer learning (using pretrained ImageNet models), gives a 2X boost on out-of-domain data vs. from scratch training Neural networks are highly effective tools for pose esti- Test (out of domain), mation. However, as in other computer vision tasks, robust- trained from scratch ness to out-of-domain data remains a challenge, especially for small training sets that are common for real-world ap- plications. Here, we probe the generalization ability with three architecture classes (MobileNetV2s, ResNets, and Ef- ficientNets) for pose estimation. We developed a dataset of 30 horses that allowed for both “within-domain” and “out- of-domain” (unseen horse) benchmarking—this is a cru- Test (out of domain), cial test for robustness that current human pose estimation w/ transfer learning benchmarks do not directly address. We show that better MobileNetV2-0.35 EfficientNet-B0 MobileNetV2-1 EfficientNet-B3 ImageNet-performing architectures perform better on both ResNet-50 Train within- and out-of-domain data if they are first pretrained Test (within domain) w/transfer learning on ImageNet. We additionally show that better ImageNet trained from scratch models generalize better across animal species. Further- more, we introduce Horse-C, a new benchmark for common Figure 1. Transfer Learning boosts performance, especially on corruptions for pose estimation, and confirm that pretrain- out-of-domain data. Normalized pose estimation error vs. Ima- ing increases performance in this domain shift context as geNet Top 1% accuracy with different backbones. -

A Primer on Motion Capture with Deep Learning: Principles, Pitfalls and Perspectives
A Primer on Motion Capture with Deep Learning: Principles, Pitfalls and Perspectives Alexander Mathis1,2,3,5,*, Steffen Schneider3,4, Jessy Lauer1,2,3, and Mackenzie W. Mathis1,2,3,* 1Center for Neuroprosthetics, Center for Intelligent Systems, Swiss Federal Institute of Technology (EPFL), Lausanne, Switzerland 2Brain Mind Institute, School of Life Sciences, Swiss Federal Institute of Technology (EPFL), Lausanne, Switzerland 3The Rowland Institute at Harvard, Harvard University, Cambridge, MA USA 4University of Tübingen & International Max Planck Research School for Intelligent Systems, Germany 5lead Contact *Corresponding authors: alexander.mathis@epfl.ch, mackenzie.mathis@epfl.ch Extracting behavioral measurements non-invasively from video other animal’s motion is determined by the geometric struc- is stymied by the fact that it is a hard computational problem. tures formed by several pendulum-like motions of the ex- Recent advances in deep learning have tremendously advanced tremities relative to a joint (6). Seminal psychophysics stud- predicting posture from videos directly, which quickly impacted ies by Johansson showed that just a few coherently moving neuroscience and biology more broadly. In this primer we re- keypoints are sufficient to be perceived as human motion (6). view the budding field of motion capture with deep learning. This empirically highlights why pose estimation is a great In particular, we will discuss the principles of those novel al- summary of such video data. Which keypoints should be ex- gorithms, highlight their potential as well as pitfalls for experi- mentalists, and provide a glimpse into the future. tracted, of course, dramatically depends on the model organ- ism and the goal of the study; e.g., many are required for dense, 3D models (12–14), while a single point can suffice for analyzing some behaviors (10). -
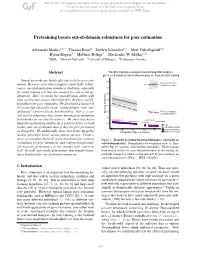
Pretraining Boosts Out-Of-Domain Robustness for Pose Estimation
Pretraining boosts out-of-domain robustness for pose estimation Alexander Mathis1,2∗† Thomas Biasi2∗ Steffen Schneider1,3 Mert Yuksekg¨ on¨ ul¨ 2,3 Byron Rogers4 Matthias Bethge3 Mackenzie W. Mathis1,2‡ 1EPFL 2Harvard University 3University of Tubingen¨ 4Performance Genetics Abstract Transfer learning (using pretrained ImageNet models), gives a 2X boost on out-of-domain data vs. from scratch training Neural networks are highly effective tools for pose esti- Test (out of domain), mation. However, as in other computer vision tasks, robust- trained from scratch ness to out-of-domain data remains a challenge, especially for small training sets that are common for real-world ap- plications. Here, we probe the generalization ability with three architecture classes (MobileNetV2s, ResNets, and Ef- ficientNets) for pose estimation. We developed a dataset of 30 horses that allowed for both “within-domain” and “out- of-domain” (unseen horse) benchmarking—this is a cru- Test (out of domain), cial test for robustness that current human pose estimation w/ transfer learning benchmarks do not directly address. We show that better MobileNetV2-0.35 EfficientNet-B0 MobileNetV2-1 EfficientNet-B3 ImageNet-performing architectures perform better on both ResNet-50 Train within- and out-of-domain data if they are first pretrained Test (within domain) w/transfer learning on ImageNet. We additionally show that better ImageNet trained from scratch models generalize better across animal species. Further- more, we introduce Horse-C, a new benchmark for common Figure 1. Transfer Learning boosts performance, especially on corruptions for pose estimation, and confirm that pretrain- out-of-domain data. Normalized pose estimation error vs. -

Download Speaker Bios
SPEAKER BIOS Lisa Federer Lisa Federer is the Data Science and Open Science Librarian at the National Library of Medicine, focusing on developing efforts to support workforce development and enhance capacity in the biomedical research and library communities for data science and open science. Prior to joining NLM, Lisa spent five years as the Research Data Informationist at the National Institutes of Health Library, where she developed and ran the Library’s Data Services Program. She holds a PhD in information studies from the University of Maryland and an MLIS from the University of California-Los Angeles, as well as graduate certificates in data science and data visualization. Her research focuses on quantifying and characterizing biomedical data reuse and development of meaningful scholarly metrics for shared data. Alexander Mathis Alexander Mathis is a Postdoctoral Fellow at Harvard University working with Prof. Venkatesh N. Murthy. He is interested in elucidating how the brain gives rise to adaptive behavior. For those purposes, he develops deep learning methods to analyze animal behavior. His PhD thesis with Prof. Andreas Herz focused on deriving properties of grid cells from optimal coding assumptions. In 2020 he will start his group at the Ecole Polytechnique Fédérale de Lausanne. Casey Greene Casey is an Associate Professor of Systems Pharmacology and Translational Therapeutics in the Perelman School of Medicine at the University of Pennsylvania. His lab develops deep learning methods that integrate distinct large-scale datasets to extract the rich and intrinsic information embedded in such integrated data. A core mission of his lab is bringing these capabilities into every molecular biology lab.