Dias Nummer 1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Product Sheet Info
Product Information Sheet for HM-289 Facklamia sp., Strain HGF4 Incubation: Temperature: 37°C Catalog No. HM-289 Atmosphere: Aerobic Propagation: 1. Keep vial frozen until ready for use, then thaw. For research use only. Not for human use. 2. Transfer the entire thawed aliquot into a single tube of broth. Contributor: 3. Use several drops of the suspension to inoculate an Thomas M. Schmidt, Professor, Department of Microbiology agar slant and/or plate. and Molecular Genetics, Michigan State University, East 4. Incubate the tube, slant and/or plate at 37°C for 48 to Lansing, Michigan, USA 168 hours. Manufacturer: Citation: BEI Resources Acknowledgment for publications should read “The following reagent was obtained through BEI Resources, NIAID, NIH as Product Description: part of the Human Microbiome Project: Facklamia sp., Strain Bacteria Classification: Aerococcaceae, Facklamia HGF4, HM-289.” Species: Facklamia sp. Strain: HGF4 Biosafety Level: 2 Original Source: Facklamia sp., strain HGF4 is a human Appropriate safety procedures should always be used with gastrointestinal isolate.1 this material. Laboratory safety is discussed in the following Comments: Facklamia sp., strain HGF4 (HMP ID 9411) is a publication: U.S. Department of Health and Human Services, reference genome for The Human Microbiome Project Public Health Service, Centers for Disease Control and (HMP). HMP is an initiative to identify and characterize Prevention, and National Institutes of Health. Biosafety in human microbial flora. The complete genome of Facklamia Microbiological and Biomedical Laboratories. 5th ed. sp., strain HGF4 is currently being sequenced at the J. Washington, DC: U.S. Government Printing Office, 2009; see Craig Venter Institute. -

The Microbiota Continuum Along the Female Reproductive Tract and Its Relation to Uterine-Related Diseases
ARTICLE DOI: 10.1038/s41467-017-00901-0 OPEN The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases Chen Chen1,2, Xiaolei Song1,3, Weixia Wei4,5, Huanzi Zhong 1,2,6, Juanjuan Dai4,5, Zhou Lan1, Fei Li1,2,3, Xinlei Yu1,2, Qiang Feng1,7, Zirong Wang1, Hailiang Xie1, Xiaomin Chen1, Chunwei Zeng1, Bo Wen1,2, Liping Zeng4,5, Hui Du4,5, Huiru Tang4,5, Changlu Xu1,8, Yan Xia1,3, Huihua Xia1,2,9, Huanming Yang1,10, Jian Wang1,10, Jun Wang1,11, Lise Madsen 1,6,12, Susanne Brix 13, Karsten Kristiansen1,6, Xun Xu1,2, Junhua Li 1,2,9,14, Ruifang Wu4,5 & Huijue Jia 1,2,9,11 Reports on bacteria detected in maternal fluids during pregnancy are typically associated with adverse consequences, and whether the female reproductive tract harbours distinct microbial communities beyond the vagina has been a matter of debate. Here we systematically sample the microbiota within the female reproductive tract in 110 women of reproductive age, and examine the nature of colonisation by 16S rRNA gene amplicon sequencing and cultivation. We find distinct microbial communities in cervical canal, uterus, fallopian tubes and perito- neal fluid, differing from that of the vagina. The results reflect a microbiota continuum along the female reproductive tract, indicative of a non-sterile environment. We also identify microbial taxa and potential functions that correlate with the menstrual cycle or are over- represented in subjects with adenomyosis or infertility due to endometriosis. The study provides insight into the nature of the vagino-uterine microbiome, and suggests that sur- veying the vaginal or cervical microbiota might be useful for detection of common diseases in the upper reproductive tract. -

Current Developments of Bacteriocins, Screening Methods and Their Application in Aquaculture and Aquatic Products
Biocatalysis and Agricultural Biotechnology 22 (2019) 101395 Contents lists available at ScienceDirect Biocatalysis and Agricultural Biotechnology journal homepage: http://www.elsevier.com/locate/bab Current developments of bacteriocins, screening methods and their application in aquaculture and aquatic products Jinsong Wang a,b,*,1, Shuwen Zhang b,1, Yujie Ouyang b, Rui Li b a Food College, Shanghai Ocean University, No.999, Huchenghuan Road, Nanhui New City, Shanghai, 201306, China b College of Bioengineering, Jingchu University of Technology, No.33, Xiangshan Road, Jingmen, 448000, China ARTICLE INFO ABSTRACT Keywords: The bacteriocins of lactic acid bacteria (LAB) is a kind of antibacterial peptide or protein synthesized by LAB in Bacteriocins vivo. They have been the focus of much research because LAB and their metabolic products are generally Lactic acid bacteria regarded as safe and have potential application in the aquaculture and aquatic products. At present, the dis Aquatic products covery of new bacteriocins involves complicated screening, identification, purification, and characterization Antimicrobial peptide processes which impact the application of these bacteriocins. This review summarizes the systematic classifi Bacteriocin applications cation of bacteriocins, especially the bacteriocins inhibiting Gram-negative and Gram-positive bacterium, and the species, genus, family and order of LAB. At the same time, the methods of screening of antimicrobial peptide production are expounded and bacteriocin applications of LAB in aquaculture and storage of aquatic products are described. All of these will provide the faster, easier, and more efficientnew antimicrobial explorations and basic knowledge for the development and applications of bacteriocins from LAB. 1. Introduction bacteria, including several aquatic products such as shellfish (Pinto, 2009), shrimp (Feliatra et al., 2018), fermented Fish (Wang et al., 2018). -

Insights Into 6S RNA in Lactic Acid Bacteria (LAB) Pablo Gabriel Cataldo1,Paulklemm2, Marietta Thüring2, Lucila Saavedra1, Elvira Maria Hebert1, Roland K
Cataldo et al. BMC Genomic Data (2021) 22:29 BMC Genomic Data https://doi.org/10.1186/s12863-021-00983-2 RESEARCH ARTICLE Open Access Insights into 6S RNA in lactic acid bacteria (LAB) Pablo Gabriel Cataldo1,PaulKlemm2, Marietta Thüring2, Lucila Saavedra1, Elvira Maria Hebert1, Roland K. Hartmann2 and Marcus Lechner2,3* Abstract Background: 6S RNA is a regulator of cellular transcription that tunes the metabolism of cells. This small non-coding RNA is found in nearly all bacteria and among the most abundant transcripts. Lactic acid bacteria (LAB) constitute a group of microorganisms with strong biotechnological relevance, often exploited as starter cultures for industrial products through fermentation. Some strains are used as probiotics while others represent potential pathogens. Occasional reports of 6S RNA within this group already indicate striking metabolic implications. A conceivable idea is that LAB with 6S RNA defects may metabolize nutrients faster, as inferred from studies of Echerichia coli.Thismay accelerate fermentation processes with the potential to reduce production costs. Similarly, elevated levels of secondary metabolites might be produced. Evidence for this possibility comes from preliminary findings regarding the production of surfactin in Bacillus subtilis, which has functions similar to those of bacteriocins. The prerequisite for its potential biotechnological utility is a general characterization of 6S RNA in LAB. Results: We provide a genomic annotation of 6S RNA throughout the Lactobacillales order. It laid the foundation for a bioinformatic characterization of common 6S RNA features. This covers secondary structures, synteny, phylogeny, and product RNA start sites. The canonical 6S RNA structure is formed by a central bulge flanked by helical arms and a template site for product RNA synthesis. -

Facklamia Sourekii Sp. Nov., Isolated F Rom 1 Human Sources
International Journal of Systematic Bacteriology (1999), 49, 635-638 Printed in Great Britain Facklamia sourekii sp. nov., isolated f rom NOTE 1 human sources Matthew D. Collins,' Roger A. Hutson,' Enevold Falsen2 and Berit Sj6den2 Author for correspondence : Matthew D. Collins. Tel : + 44 1 18 935 7226. Fax : + 44 1 18 935 7222. e-mail : [email protected] 1 Department of Food Two strains of a Gram-positive catalase-negative, facultatively anaerobic Science and Technology, coccus originating from human sources were characterized by phenotypic and University of Reading, Whiteknights, molecular taxonomic methods. The strains were found to be identical to each Reading RG6 6AP, other based on 165 rRNA gene sequencing and constitute a new subline within UK the genus Facklamia. The unknown bacterium was readily distinguished from * Culture Collection, Facklamis hominis and Facklamia ignava by biochemicaltests and Department of Clinical electrophoretic analysis of whole-cell proteins. Based on phylogenetic and Bacteriology, University of Goteborg, Sweden phenotypic evidence it is proposed that the unknown bacterium be classified as Facklamia sourekii sp. nov., the type strain of which is CCUG 28783AT. Keywords : Facklamia sourekii, taxonomy, phylogeny, 16s rRNA The Gram-positive catalase-negative cocci constitute a Gram-positive coccus-shaped organisms, which con- phenotypically heterogeneous group of organisms stitute a third species of the genus Facklamia, Fack- which belong to the Clostridium branch of the Gram- lamia sourekii sp. nov. This report adds to the many positive bacteria. This broad group of organisms new taxa and the diversity of Gram-positive catalase- includes many human and animal pathogens (e.g. -

Individual Differences in Members of Actinobacteria, Bacteroidetes, and Firmicutes Is
bioRxiv preprint doi: https://doi.org/10.1101/2021.07.23.453592; this version posted July 24, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Individual differences in members of Actinobacteria, Bacteroidetes, and Firmicutes is associated with resistance or vulnerability to addiction-like behaviors in heterogeneous stock rats 1 1 1 1 1 1 1 S. Simpson , G. de Guglielmo , M. Brennan , L. Maturin , G. Peters , H. Jia , E. Wellmeyer , S. Andrews1, L. Solberg Woods2, A. A. Palmer1,3, O. George1* 1Department of Psychiatry, University of California San Diego, La Jolla CA 92093, USA 2Department of Internal MediCine, SeCtion on MoleCular MediCine, Wake Forest University SChool of MediCine, Winston-Salem, NC 27157 3 Institute for GenomiC MediCine, University of California, San Diego, La Jolla, CA 92093 *Corresponding Author Dr. Olivier George, Department of Psychiatry University of California San Diego La Jolla, CA 92093, USA. Tel: +18582465538. E-mail:[email protected] bioRxiv preprint doi: https://doi.org/10.1101/2021.07.23.453592; this version posted July 24, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract An emerging element in psychiatry is the gut-brain-axis, the bi-direCtional CommuniCation pathways between the gut miCrobiome and the brain. A prominent hypothesis, mostly based on preCliniCal studies, is that individual differences in the gut miCrobiome Composition and drug- induced dysbiosis may be associated with vulnerability to psychiatriC disorders including substance use disorder. -

Microbial Interactions Within the Cheese Ecosystem and Their Application to Improve Quality and Safety
foods Review Microbial Interactions within the Cheese Ecosystem and Their Application to Improve Quality and Safety Baltasar Mayo * , Javier Rodríguez, Lucía Vázquez and Ana Belén Flórez Departamento de Microbiología y Bioquímica, Instituto de Productos Lácteos de Asturias (IPLA), Consejo Superior de Investigaciones Científicas (CSIC), Paseo Río Linares s/n, 33300 Villaviciosa, Spain; [email protected] (J.R.); [email protected] (L.V.); abfl[email protected] (A.B.F.) * Correspondence: [email protected]; Tel.: +34-985893345 Abstract: The cheese microbiota comprises a consortium of prokaryotic, eukaryotic and viral popula- tions, among which lactic acid bacteria (LAB) are majority components with a prominent role during manufacturing and ripening. The assortment, numbers and proportions of LAB and other microbial biotypes making up the microbiota of cheese are affected by a range of biotic and abiotic factors. Cooperative and competitive interactions between distinct members of the microbiota may occur, with rheological, organoleptic and safety implications for ripened cheese. However, the mechanistic details of these interactions, and their functional consequences, are largely unknown. Acquiring such knowledge is important if we are to predict when fermentations will be successful and understand the causes of technological failures. The experimental use of “synthetic” microbial communities might help throw light on the dynamics of different cheese microbiota components and the interplay Citation: Mayo, B.; Rodríguez, J.; between them. Although synthetic communities cannot reproduce entirely the natural microbial di- Vázquez, L.; Flórez, A.B. Microbial versity in cheese, they could help reveal basic principles governing the interactions between microbial Interactions within the Cheese types and perhaps allow multi-species microbial communities to be developed as functional starters. -

Effects of the Inclusion of Fermented Mulberry Leaves and Branches in the Gestational Diet on the Performance and Gut Microbiota of Sows and Their Offspring
microorganisms Article Effects of the Inclusion of Fermented Mulberry Leaves and Branches in the Gestational Diet on the Performance and Gut Microbiota of Sows and Their Offspring Yuping Zhang 1,2 , Chang Yin 1 , Martine Schroyen 2, Nadia Everaert 2, Teng Ma 1,* and Hongfu Zhang 1 1 State Key Laboratory of Animal Nutrition, Institute of Animal Sciences, Chinese Academy of Agricultural Sciences, Beijing 100193, China; [email protected] (Y.Z.); [email protected] (C.Y.); [email protected] (H.Z.) 2 Precision Livestock and Nutrition Laboratory, TERRA Teaching and Research Centre, Gembloux Agro-Bio Tech, University of Liège, 5030 Gembloux, Belgium; [email protected] (M.S.); [email protected] (N.E.) * Correspondence: [email protected]; Tel.: +86-130-2198-3772 Abstract: Fermented feed mulberry (FFM), being rich in dietary fiber, has not been fully evaluated to be used in sow’s diet. In this study, we investigated the effects of 25.5% FFM supplemented in gestation diets on the performance and gut microbiota of sows and their offspring. Results showed that the serum concentration of glucose, progesterone, and estradiol were not affected by the dietary treatment, while the level of serum insulin and fecal short chain fatty acid were both reduced in FFM group on gestation day 60 (G60, p < 0.05). Additionally, FFM increased both voluntary feed intake and Citation: Zhang, Y.; Yin, C.; weaning litter weight (p < 0.05), while decreased the losses of both Backfat thickness and bodyweight Schroyen, M.; Everaert, N.; Ma, T.; throughout lactation (p < 0.05). 16S rRNA sequencing showed FFM supplementation significantly Zhang, H. -

Co-Infection in Critically Ill Patients with COVID-19: an Observational Cohort Study from England
medRxiv preprint doi: https://doi.org/10.1101/2020.10.27.20219097; this version posted October 27, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. It is made available under a CC-BY-NC-ND 4.0 International license . Co-infection in COVID-19 Co-infection in critically ill patients with COVID-19: An observational cohort study from England Vadsala Baskaran1,2,3, Hannah Lawrence1,2,3, Louise Lansbury2, Karmel Webb2, Shahideh Safavi3,4, Izzah Zainuddin1, Tausif Huq1, Charlotte Eggleston1, Jayne Ellis5, Clare Thakker5, Bethan Charles6, Sara Boyd8,9, Tom Williams8, Claire Phillips10, Ethan Redmore10, Sarah Platt11, Eve Hamilton11, Andrew Barr11 , Lucy Venyo11, Peter Wilson5, Tom Bewick12, Priya Daniel12, Paul Dark6,7, Adam R Jeans6, Jamie McCanny8, Jonathan D Edgeworth8, Martin J Llewelyn10, Matthias L Schmid11, Tricia M McKeever2,3, Martin Beed13,14, Wei Shen Lim1,3 Running Title: Co-infection in COVID-19 Institutions: 1 Department of Respiratory Medicine, Nottingham University Hospital NHS Trust, Nottingham NG5 1PB, UK 2 Division of Epidemiology and Public Health, School of Medicine, University of Nottingham, Clinical Sciences Building, Nottingham City Hospital Campus, Hucknall Road, Nottingham NG5 1PB, UK 3 NIHR Nottingham Biomedical Research Centre, Queen’s Medical Centre, Nottingham NG7 2UH, UK 4 Division of Respiratory Medicine, School of Medicine, University of Nottingham, Queens Medical Centre, Derby Rd, -

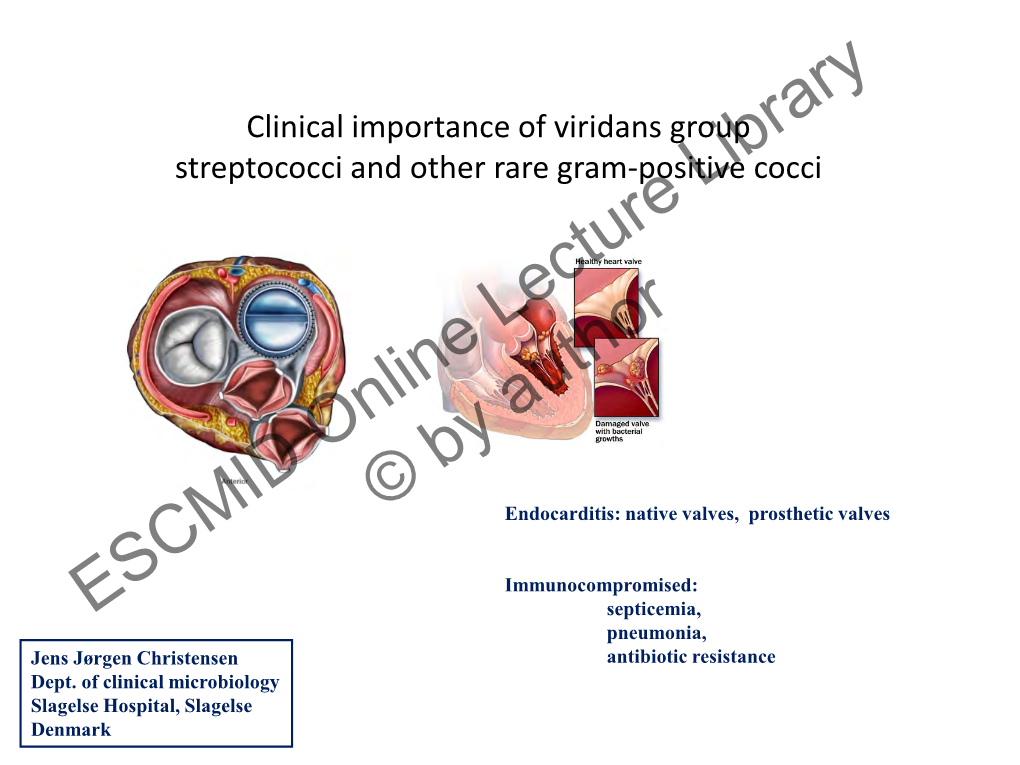
Streptococcus Laboratory General Methods
The reference used for compiling the methods in Section I is: Murray, P.R., Baron, E. J., Jorgensen, J.J., Pfaller, M.A., and Yolken, R.H. Manual of Clinical Microbiology, 8th ed. ASM Press: Washington, DC, 2003. The Streptococcus species identification methods in Section II were compiled by Dr. Lynn Shewmaker. Also thanks to input from several individuals, including Richard Facklam and Lucia Teixeira. Section I. 1. Accuprobe-Enterococcus Test………….………..4 2. Accuprobe-Pneumococcus Test …………..….….4 3. Acid formation in carbohydrate broth..................5 4. Arginine Hydrolysis……………………….……….6 5. Bacitracin Test……………………………………..7 6. Bile-esculin Test…………………………………...8 7. Bile solubility Test …………………………………9 8. CAMP Test…......................................................10 9. Catalase Test......................................................11 10. Clindamycin test………………………………….12 11. Esculin hydrolysis……………………………….. 13 12. Gas from MRS broth……………………………...14 13. Gram Stain………………………………………...15 14. Growth at 10 & 45C……………………………. 17 15. Hemolysis………………………………………….18 16. Hippurate hydrolysis…………………………… 19 17. Lancefield Group Antigen………………………..20 18. Leucine amino peptidase (LAP)…………………21 19. Litmus Milk Test…………………………………..22 20. Motility………………………………………………23 21. 6.5% NaCl Tolerace Test...................................24 22. Optochin…………………………………………….25 23. Pigmentation....................................................... 26 24. Pyridoxal Requirement Test (Vitamin B6)……….27 25. Pyrrolidonlarylamindase (PYR)............................28 26. -
A First Record of Facklamia Hominis
Plant Archives Vol. 19 No. 2, 2019 pp. 4294-4298 e-ISSN:2581-6063 (online), ISSN:0972-5210 A FIRST RECORD OF FACKLAMIA HOMINIS ISOLATED FROM COMMON CARP (CYPRINUS CARPIO) CULTIVATED IN FLOATING CAGES AT AL-HILLA RIVER, BABYLON PROVINCE: ANTIBIOTIC RESISTANT AND SUSCEPTIBILITY OF PLANT SPECIES Sadeq M. Al-Haider1, Khalidah S. Al-Niaeem2* and Amjed K. Resen2 1College of Veterinary Medicine, University of Al-Qasim Green, Iraq. 2*Department of Fisheries and Marine Resources, College of Agriculture, University of Basrah, Basrah, Iraq. Abstract This study was carried out to investigate the occurrence of potentially pathogenic species of Gram-positive bacterium in common carp cultivated in floating cages at Al-Hilla river. A total of 144 fishes were collected from fish farms during the period December 2017- November 2018. Bacteria were identified using the VITEK 2 system and selected biochemical tests. Here we report the first fish’s case of gills due to F. hominis, found in common carp cultivated in floating cages at Al-Hilla river. Antibiotic susceptibility (17 antimicrobial) was using the VITEK 2 system. Significant sensitive was produced only by Ciprofloxacin, Ertapenem and Levofloxacin, when tested in vitro. The plant species susceptibility test was conducted using disc diffusion method on blood agar. Procedure was based on the standardized disc agar diffusion method for antimicrobial susceptibility tests. After incubation, the diameter of the zone of inhibition was measured to determine the sensitivity of the six plant species extracted with alcohol in four different concentrations (10, 15, 20 and 25%). The potential of plant extracts to inhibit the infection caused by F. -

A Diet High in Sugar and Fat Influences Neurotransmitter Metabolism And
Guo et al. Translational Psychiatry (2021) 11:328 https://doi.org/10.1038/s41398-021-01443-2 Translational Psychiatry ARTICLE Open Access A diet high in sugar and fat influences neurotransmitter metabolism and then affects brainfunctionbyalteringthegutmicrobiota Yinrui Guo 1,XiangxiangZhu2,3,MiaoZeng2,4,LongkaiQi2, Xiaocui Tang2, Dongdong Wang2,MeiZhang4, Yizhen Xie2, Hongye Li3,XinYang5 and Diling Chen 2 Abstract Gut microbiota (GM) metabolites can modulate the physiology of the host brain through the gut–brain axis. We wished to discover connections between the GM, neurotransmitters, and brain function using direct and indirect methods. A diet with increased amounts of sugar and fat (high-sugar and high-fat (HSHF) diet) was employed to disturb the host GM. Then, we monitored the effect on pathology, neurotransmitter metabolism, transcription, and brain circularRNAs (circRNAs) profiles in mice. Administration of a HSHF diet-induced dysbacteriosis, damaged the intestinal tract, changed the neurotransmitter metabolism in the intestine and brain, and then caused changes in brain function and circRNA profiles. The GM byproduct trimethylamine-n-oxide could degrade some circRNAs. The basal level of the GM decided the conversion rate of choline to trimethylamine-n-oxide. A change in the abundance of a single bacterial strain could influence neurotransmitter secretion. These findings suggest that a new link between metabolism, brain circRNAs, and GM. Our data could enlarge the “microbiome–transcriptome” linkage library and provide more information on the gut–brain axis. Hence, our findings could provide more information on the interplay fi 1234567890():,; 1234567890():,; 1234567890():,; 1234567890():,; between the gut and brain to aid the identi cation of potential therapeutic markers and mechanistic solutions to complex problems encountered in studies of pathology, toxicology, diet, and nutrition development.