Downloaded L120 Sdf Files Were Manually Curated, Merged Into One Sdf File, and Duplicates Eliminated 63 F 139 Using Open Babel Vs 2.3.1
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

In Search of a Protein Nucleator of Hydroxyapatite in Bone
IN SEARCH OF A PROTEIN NUCLEATOR OF HYDROXYAPATITE IN BONE Carmel O Domeni cucci A rhesis subrnitted in conformity with the requirements for the degree of Doctor of Philosophy Graduate Department of Biochemistq University of Toronto O Copyright by Canne10 Domenicucci 1997 National Library Bibiiothéque nationale B*m of Canada du Canada Acquisitions and Acquisitions et Bibliographic Services sewices bibliographiques 395 Wellington Street 395. rue Wellington OttawaON K1AON4 OrtawaON KIAON4 Canada Canada Your iVe Vaire rehrefue Our !W Notre referwice The author has granted a non- L'auteur a accordé une licence non exclusive licence allowing the exclusive permettant à la National Library of Canada to Bibliothèque nationale du Canada de reproduce, loan, distribute or seil reproduire, prêter, distribuer ou copies of ths thesis in rnicroform, vendre des copies de cette thèse sous paper or electronic formats. la forme de microfiche/fh, de reproduction sur papier ou sur format électronique. The author retallis ownership of the L'auteur conserve la propriété du copyright in ths thesis. Neither the droit d'auteur qui protège cette thése. thesis nor substantial extracts &om it Ni la thèse ni des extraits substantiels may be printed or otherwise de celle-ci ne doivent être imprimés reproduced without the author's ou autrement reproduits sans son permission. autorisation. Yn Search of a Protein Nucleator of Hydroxyapatite in Bone" Doctor of Philosophy. 1997 Carme10 Domenicucci Department of Biochemistry University of Toronto The formation of mineralized conneciive tissues is characterized by the nucleation of hydroxyapatite crystals that are generated initiaily within the gap region of collagen fibrils. However, the mechanisrn of mineral nuckation has not been resolved. -

VIEW Open Access Vitamin K Antagonist Use: Evidence of the Difficulty of Achieving and Maintaining Target INR Range and Subsequent Consequences Jeff R
Schein et al. Thrombosis Journal (2016) 14:14 DOI 10.1186/s12959-016-0088-y REVIEW Open Access Vitamin K antagonist use: evidence of the difficulty of achieving and maintaining target INR range and subsequent consequences Jeff R. Schein1, C. Michael White2,3, Winnie W. Nelson1, Jeffrey Kluger3, Elizabeth S. Mearns2 and Craig I. Coleman2,3* Abstract Vitamin K antagonists (VKAs) are effective oral anticoagulants that are titrated to a narrow therapeutic international normalized ratio (INR) range. We reviewed published literature assessing the impact of INR stability - getting into and staying in target INR range - on outcomes including thrombotic events, major bleeding, and treatment costs, as well as key factors that impact INR stability. A time in therapeutic range (TTR) of ≥65 % is commonly accepted as the definition of INR stability. In the real-world setting, this is seldom achieved with standard-of-care management, thus increasing the patients’ risks of thrombotic or major bleeding events. There are many factors associated with poor INR control. Being treated in community settings, newly initiated on a VKA, younger in age, or nonadherent to therapy, as well as having polymorphisms of CYP2C9 or VKORC1, or multiple physical or mental co-morbid disease states have been associated with lower TTR. Clinical prediction tools are available, though they can only explain <10 % of the variance behind poor INR control. Clinicians caring for patients who require anticoagulation are encouraged to intensify diligence in INR management when using VKAs and to consider appropriate use of newer anticoagulants as a therapeutic option. Keywords: Vitamin K antagonists, International normalized ratio, Anticoagulation, Atrial fibrillation, Venous thromboembolism Background (within 72–96 h after dosing) and an anticoagulated Vitamin K antagonists (VKAs) such as warfarin inhibit state. -

Xerox University Microfilms 300 North Ze«B Road Ann Arbor, Michigan 48106 75-11,067
ASYMMETRIC STRUCTURE OF PROTHROMBIN (FACTOR II): CALCIUM AND LIPID BINDING SITES, AND CARBOHYDRATE DISTRIBUTION Item Type text; Dissertation-Reproduction (electronic) Authors Benson, Bradley Jonnell, 1945- Publisher The University of Arizona. Rights Copyright © is held by the author. Digital access to this material is made possible by the University Libraries, University of Arizona. Further transmission, reproduction or presentation (such as public display or performance) of protected items is prohibited except with permission of the author. Download date 10/10/2021 08:15:19 Link to Item http://hdl.handle.net/10150/290366 INFORMATION TO USERS This material was produced from a microfilm copy of the original document. While the most advanced technological means to photograph and reproduce this document have been used, the quality is heavily dependent upon the quality of the original submitted. The following explanation of techniques is provided to help you understand markings or patterns which may appear on this reproduction. 1. The sign or "target" for pages apparently lacking from the document photographed is "Missing Page(s)". If it was possible to obtain the missing page(s) or section, they are spliced into the film along with adjacent pages. This may have necessitated cutting thru an image and duplicating adjacent pages to insure you complete continuity. 2. When an image on the film is obliterated with a large round black mark, it is an indication that the photographer suspected that the copy may have moved during exposure and thus cause a blurred image. You will find a good image of the page in the adjacent frame. 3. -

VITAMIN K SUBSTANCES 1. Exposure Data
VITAMIN K SUBSTANCES Vitamin K comprises a group of substances, which are widespread in nature and are an essential co-factor in humans in the synthesis of several proteins that play a role in haemostasis and others that may be important in calcium homeostasis. The K vitamins all contain the 2-methyl-1,4-naphthoquinone (menadione) moiety, and the various naturally occurring forms differ in the alkyl substituent at the 3-position. Phylloquinone (vitamin K1) is 2-methyl-3-phytyl-1,4-naphthoquinone and is widely found in higher plants, including green leafy vegetables, and in green and blue algae. The menaquinones (formerly vitamin K2) have polyisoprenyl substituents at the 3-position and are produced by bacteria. The compound menadione (formerly vitamin K3) lacks an alkyl group at the 3-position but can be alkylated in vivo in some species. Several synthetic water-soluble derivatives, such as the sodium diphosphate ester of menadiol and the addition product of menadione with sodium bisulfite, also have commercial applications (National Research Council, 1989; Gennaro, 1995; Weber & Rüttimann, 1996). 1. Exposure Data 1.1 Chemical and physical data 1.1.1 Nomenclature, structural and molecular formulae and relative molecular masses Vitamin K (generic) Chem. Abstr. Serv. Reg. No.: 12001-79-5 Chem. Abstr. Name: Vitamin K Vitamin K1 (generic) Chem. Abstr. Serv. Reg. No.: 11104-38-4 Chem. Abstr. Name: Vitamin K1 Phylloquinone Chem. Abstr. Serv. Reg. No.: 84-80-0 Deleted CAS Reg. Nos.: 10485-69-5; 15973-57-6; 50926-17-5 –417– 418 IARC MONOGRAPHS -

Downloaded From
Towards improvement of oral anticoagulant therapy Leeuwen, Y. van Citation Leeuwen, Y. van. (2009, April 2). Towards improvement of oral anticoagulant therapy. Retrieved from https://hdl.handle.net/1887/13716 Version: Corrected Publisher’s Version Licence agreement concerning inclusion of doctoral License: thesis in the Institutional Repository of the University of Leiden Downloaded from: https://hdl.handle.net/1887/13716 Note: To cite this publication please use the final published version (if applicable). CHAPTER 7 A Randomised Comparison of the Quality of anticoagulant therapy with Two Vitamin K antagonists: Warfarin versus Phenprocoumon Van Leeuwen Y, Gebuis EPA, van der Meer FJM, Rosendaal FR 118 Chapter 7 Abstract Context Several studies have compared different vitamin K antagonists with regard to the quality of treatment, e.g. stability. The results were mostly, but not always, in favour of the longer acting vitamin K antagonists. Only one randomised study compared warfarin and phenprocoumon. Objective The aim of our study was to investigate whether oral anticoagulant treatment with warfarin leads to a more or less stable anticoagulant status than is achieved with phenprocoumon. Methods We conducted a randomised controlled trial among patients with an indication for anticoagulant treatment at the Leiden Anticoagulation Clinic. We included patients who initiated oral anticoagulant therapy and patients who were already using a different vitamin K antagonist (acenocoumarol), and who were switched. We compared an oral anticoagulant treatment with warfarin with a treatment with phenprocoumon. Follow-up was 6 months. The primary outcome measure was the percentage of time spent in therapeutic range. Results From March 1st 2004 to January 2008, 312 patients finished the follow period and were included in this analysis. -

A Specific Antidote for Reversal of Anticoagulation by Direct and Indirect Inhibitors of Coagulation Factor Xa
ARTICLES A specific antidote for reversal of anticoagulation by direct and indirect inhibitors of coagulation factor Xa Genmin Lu1, Francis R DeGuzman2, Stanley J Hollenbach2, Mark J Karbarz1, Keith Abe2, Gail Lee2, Peng Luan1, Athiwat Hutchaleelaha3, Mayuko Inagaki3, Pamela B Conley1, David R Phillips1 & Uma Sinha1 Inhibitors of coagulation factor Xa (fXa) have emerged as a new class of antithrombotics but lack effective antidotes for patients experiencing serious bleeding. We designed and expressed a modified form of fXa as an antidote for fXa inhibitors. This recombinant protein (r-Antidote, PRT064445) is catalytically inactive and lacks the membrane-binding g-carboxyglutamic acid domain of native fXa but retains the ability of native fXa to bind direct fXa inhibitors as well as low molecular weight heparin– activated antithrombin III (ATIII). r-Antidote dose-dependently reversed the inhibition of fXa by direct fXa inhibitors and corrected the prolongation of ex vivo clotting times by such inhibitors. In rabbits treated with the direct fXa inhibitor rivaroxaban, r-Antidote restored hemostasis in a liver laceration model. The effect of r-Antidote was mediated by reducing plasma anti-fXa activity and the non–protein bound fraction of the fXa inhibitor in plasma. In rats, r-Antidote administration dose-dependently and completely corrected increases in blood loss resulting from ATIII-dependent anticoagulation by enoxaparin or fondaparinux. r-Antidote has the potential to be used as a universal antidote for a broad range of fXa inhibitors. Warfarin, a vitamin K antagonist, is widely used as an anticoagulant As direct oral fXa inhibitors such as rivaroxaban and apixaban have for the prevention and treatment of thrombotic diseases. -

WARFARIN Class: Vitamin K Antagonist Indications : Prophylaxis
WARFARIN Class: Vitamin K Antagonist Indications : Prophylaxis and treatment of thromboembolic disorders (eg, venous, pulmonary) and embolic complications arising from atrial fibrillation or cardiac valve replacement; adjunct to reduce risk of systemic embolism (eg, recurrent MI, stroke) after myocardial infarction. Unlabeled: Prevention of recurrent transient ischemic attacks Available dosage form in the hospital: 3MG TAB, 5MG TAB Dosage: Note: Labeling identifies genetic factors which may increase patient sensitivity to warfarin. Specifically, genetic variations in the proteins CYP2C9 and VKORC1, responsible for warfarin’s primary metabolism and pharmacodynamic activity, respectively, have been identified as predisposing factors associated with decreased dose requirement and increased bleeding risk. Genotyping tests are available, and may provide guidance on initiation of anticoagulant therapy. The American College of Chest Physicians recommends against the use of routine pharmacogenomic testing to guide dosing (Guyatt, 2012). For management of elevated INRs as a result of warfarin therapy, see Additional Information/Pharmacotherapy Pearls for guidance. -Prevention/treatment of thrombosis/embolism: -I.V. (administer as a slow bolus injection): 2-5 mg/day -Oral: Initial dosing must be individualized. Consider the patient (hepatic function, cardiac function, age, nutritional status, concurrent therapy, risk of bleeding) in addition to prior dose response (if available) and the clinical situation. Start 2-5 mg once daily for 2 days or for healthy individuals, 10 mg once daily for 2 days; lower doses (eg, 5 mg once daily) recommended for patients with confirmed HIT once platelet recovery has occurred.In patients with acute venous thromboembolism, initiation may begin on the first or second day of low molecular weight heparin or unfractionated heparin therapy.Adjust dose according to INR results; usual maintenance dose ranges from 2-10 mg daily (individual patients may require loading and maintenance doses outside these general guidelines). -

Study Protocol
<Eliquis> <B0661120> Safety and effectiveness evaluation of patients with non-valvular atrial fibrillation treated with OACs: Comparison between NOACs and warfarin (CER3) <Final ver.1.1><20180508> NON-INTERVENTIONAL (NI) STUDY PROTOCOL Revision Effective Date Summary of Revisions 1.1 2018/5/08 Schedule(Milestone) has been revised 090177e191faa505\Approved\Approved On: 01-Oct-2019 06:39 (GMT) Page 1 of 16 <Eliquis> <B0661120> Safety and effectiveness evaluation of patients with non-valvular atrial fibrillation treated with OACs: Comparison between NOACs and warfarin (CER3) <Final ver.1.1><20180508> Study information Safety and effectiveness evaluation of patients with non- Title valvular atrial fibrillation treated with OACs: Comparison between NOACs and warfarin (CER3) Protocol number B0661120 Protocol version identifier 1.1 Date of last version of protocol 08MAY2018 EU Post Authorisation Study (PAS) Not applicable (this study is not PASS) register number B : Blood and blood forming organs B01: Antithrombotic agents Active substance B01A : Antithrombotic agents B01AF: Direct factor Xa inhibitors B01AF02: Apixaban Medicinal product Eliquis (Apixaban) The research questions are: 1) is there any difference in the risk of major bleeding and composite of ischemic stroke, hemorrhagic stroke or systemic embolism (stroke/SE) between patients treated with warfarin and those treated with one of NOACs (apixaban, dabigatran, edoxaban or Research question and objectives rivaroxaban) in OAC naïve NVAF patients who start treatment with OACs. The primary objective is to compare the risk of major bleeding and stroke/SE in warfarin- apixaban matched cohorts, warfarin-dabigatran matched cohorts, warfarin-edoxaban matched cohorts and warfarin- rivaroxaban matched cohorts. PPD , PPD Author PPD 090177e191faa505\Approved\Approved On: 01-Oct-2019 06:39 (GMT) Page 2 of 16 <Eliquis> <B0661120> Safety and effectiveness evaluation of patients with non-valvular atrial fibrillation treated with OACs: Comparison between NOACs and warfarin (CER3) <Final ver.1.1><20180508> TABLE OF CONTENTS 1. -
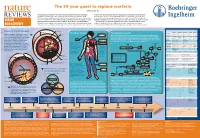
The 50-Year Quest to Replace Warfarin Jeffrey Weitz
The 50-year quest to replace warfarin Jeffrey Weitz Anticoagulants are used for the prevention and treatment of venous and arterial to check that the appropriate level of anticoagulation is reached and maintained in thrombosis, the leading cause of morbidity and mortality in the Western world. patients receiving warfarin. Consequently, warfarin is underused, and the level of Warfarin — the prototype oral anticoagulant — is a vitamin K antagonist that has been anticoagulation is often suboptimal even when it is administered. These drawbacks in clinical use since the 1950s. Although they are effective, vitamin K antagonists have highlight the need for new oral anticoagulants. The first drug to be approved — in 2010 DRUG several drawbacks, the most notable of which is the propensity to cause bleeding. — as an alternative to warfarin was dabigatran, a direct thrombin inhibitor. Clinical Other limitations include a slow onset of action, interactions with multiple other drugs studies of the direct factor Xa inhibitors rivaroxaban and apixaban have been DISCOVERY and interpatient variability in drug response. As a result, regular monitoring is required completed and could form the basis for regulatory approval as alternatives to warfarin. Comparative pharmacology of new oral anticoagulants compared to warfarin Venous and arterial thrombosis 'PFQVJGNKWO 8GUUGNYCNN Thrombotic disorders The coagulation cascade Warfarin Dabigatran Rivaroxaban Apixaban Edoxaban Arterial thrombosis. The primary trigger of Prodrug No Yes No No No (KDTKP Pulmonary This is classically divided into three pathways. The extrinsic pathway (also known as the Bioavailability Over 90% 6% 80% 60% 50% this is rupture of an atherosclerotic plaque, embolism ▶ tissue factor pathway) initiates coagulation. -

Rivaroxaban Versus Vitamin K Antagonist in Antiphospholipid
Annals of Internal Medicine ORIGINAL RESEARCH Rivaroxaban Versus Vitamin K Antagonist in Antiphospholipid Syndrome A Randomized Noninferiority Trial Josep Ordi-Ros, MD, PhD; Luis Sa´ ez-Comet, MD, PhD; Mercedes Pe´ rez-Conesa, MD; Xavier Vidal, MD, PhD; Antoni Riera-Mestre, MD, PhD; Antoni Castro-Salomo´ , MD, PhD; Jordi Cuquet-Pedragosa, MD; Vera Ortiz-Santamaria, MD; Montserrat Mauri-Plana, MD, PhD; Cristina Sole´ , PhD; and Josefina Corte´ s-Herna´ ndez, MD, PhD Background: The potential role of new oral anticoagulants in Results: After 3 years of follow-up, recurrent thrombosis oc- antiphospholipid antibody syndrome (APS) remains uncertain. curred in 11 patients (11.6%) in the rivaroxaban group and 6 (6.3%) in the VKA group (RR in the rivaroxaban group, 1.83 [95% Objective: To determine whether rivaroxaban is noninferior to CI, 0.71 to 4.76]). Stroke occurred more commonly in patients dose-adjusted vitamin K antagonists (VKAs) for thrombotic APS. receiving rivaroxaban (9 events) than in those receiving VKAs (0 Design: 3-year, open-label, randomized noninferiority trial. (EU events) (corrected RR, 19.00 [CI, 1.12 to 321.9]). Major bleeding Clinical Trials Register: EUDRA [European Union Drug Regula- occurred in 6 patients (6.3%) in the rivaroxaban group and 7 tory Authorities] code 2010-019764-36) (7.4%) in the VKA group (RR, 0.86 [CI, 0.30 to 2.46]). Post hoc analysis suggested an increased risk for recurrent thrombosis in Setting: 6 university hospitals in Spain. rivaroxaban-treated patients with previous arterial thrombosis, livedo racemosa, or APS-related cardiac valvular disease. Participants: 190 adults (aged 18 to 75 years) with thrombotic APS. -

Warfarin, a Comprehensive Review by Jay Patel – Presentation.Pdf
Warfarin: A Comprehensive Review Jay M Patel, PharmD Objectives • Explain the important pharmacodynamic and pharmacokinetic properties of warfarin • Describe the role of warfarin in the inpatient setting • Identify factors that may contribute to variability in the INR with warfarin use • Discuss warfarin initiation and dose adjustments based on a comprehensive review of the patient Disclaimer Dr. Patel has no actual or potential conflict of interest associated with this presentation Coumadin® (Warfarin) • Clinical Pharmacology – Inhibit synthesis of Vitamin K dependent clotting factors • II, VII, IX and X • Protein C and S • Mechanism of Action – “Vitamin K antagonist” (VKA) – Inhibit C1 subunit of vitamin K epoxide reductase (VKORC1) enzyme complex Coumadin® (Warfarin) • Elimination half-lives of vitamin K- dependent proteins Factor Half-Life II 42-72 hours VII 4-6 hours IX 21-30 hours X 27-48 hours Protein C 8 hours Protein S 60 hours Clotting Cascade.http://medlibes.com/uploads/Screen%20shot%202010-07-30%20at%2012.56.57%20PM.png. Accessed May 15, 2015. Warfarin Response. El Camino Hospital. http://elcaminogmi.dnadirect.com/img/content/tests/drug_response/howWarfarinAffectsBloodClotting.gif. Date Accessed May 14, 2015. Accessed May 14, 2015 Question 1 • Warfarin, through the inhibition of VKOR, inhibits the production of which factors? A. II, VIII, XII, Protein C and Protein S B. I, II, III, and X C. XI, Protein C and Protein S D. II, VII, IX, X, Protein C and Protein S Pharmacokinetics • Racemic mixture of R- and S- enantiomers – S- -

Antithrombotic Reversal – Adult – Inpatient Clinical Practice Guideline
Antithrombotic Reversal – Adult – Inpatient Clinical Practice Guideline Note: Active Table of Contents – Click to follow link EXECUTIVE SUMMARY ........................................................................................................... 3 SCOPE ................................................................................................................................... 4 METHODOLOGY .................................................................................................................... 4 DEFINITIONS.......................................................................................................................... 5 INTRODUCTION ..................................................................................................................... 5 RECOMMENDATIONS ............................................................................................................ 6 UW HEALTH IMPLEMENTATION ........................................................................................... 14 APPENDIX A. EVIDENCE GRADING SCHEME(S) ...................................................................... 15 APPENDIX B. SUMMARY OF INTERIM REVISIONS (AS APPROPRIATE) .. ERROR! BOOKMARK NOT DEFINED. REFERENCES ........................................................................................................................ 21 1 Contact for Content: Name: Anne Rose, PharmD – Pharmacy Department Phone Number: (608) 263-9738 Email Address: [email protected] CPG Contact for Changes: Name: Philip Trapskin, PharmD,