Codon Usage Bias and Peptide Properties of Pseudomonas Balearica DSM 6083T
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Doctoral Thesis 2016 UNDERSTANDING the AROMATIC HYDROCARBON DEGRADATION POTENTIAL of PSEUDOMONAS STUTZERI
Doctoral Thesis 2016 UNDERSTANDING THE AROMATIC HYDROCARBON DEGRADATION POTENTIAL OF PSEUDOMONAS STUTZERI: A PROTEO-GENOMIC APPROACH Isabel Brunet Galmés Doctoral Thesis 2016 Doctoral Program of “Microbiologia Ambiental i Biotecnologia” UNDERSTANDING THE AROMATIC HYDROCARBON DEGRADATION POTENTIAL OF PSEUDOMONAS STUTZERI: A PROTEO-GENOMIC APPROACH Isabel Brunet Galmés Thesis Supervisor: Dr. Rafael Bosch Thesis Supervisor: Dra. Balbina Nogales Doctor by the Universitat de les Illes Balears A mumpare i a mumare Agraïments Gràcies Rafel i Balbina per dirigir aquesta tesis, que és tant meva com vostre. A en Rafel per l’oportunitat que em vares donar, ja fa 7 anys, d’entrar al laboratori. Així com també per engrescar-me dins el món de la ciència, i ensenyar-me a treballar tant dins com fora del laboratori. I a na Balbina, pels mil consells que m’ha donat aquests anys, per ensenyar-me a ser més meticulosa amb el que faig i per ajudar-me amb tot el que ha pogut. A en Jordi i n’Elena, per acollir-me dins aquest grup de recerca, pels vostres consells i noves idees per continuar aquesta feina. Voldria agrair també a en Toni Bennasar i en Sebastià les crítiques constructives que m’heu anat fent al llarg d’aquests anys, des del projecte final de màster fins ara. Gràcies també a tots els companys de laboratori, a més de companys sou uns grans amics. Sempre estaré agraïda a na Marga, en Toni Busquets i n’Arantxa, pels grans consells que m’heu donat, tant dins com fora del laboratori, i per estar sempre disposats a donar-me una mà. -

Comparative Genomic Analysis of Three Pseudomonas
microorganisms Article Comparative Genomic Analysis of Three Pseudomonas Species Isolated from the Eastern Oyster (Crassostrea virginica) Tissues, Mantle Fluid, and the Overlying Estuarine Water Column Ashish Pathak 1, Paul Stothard 2 and Ashvini Chauhan 1,* 1 Environmental Biotechnology Laboratory, School of the Environment, 1515 S. Martin Luther King Jr. Blvd., Suite 305B, FSH Science Research Center, Florida A&M University, Tallahassee, FL 32307, USA; [email protected] 2 Department of Agricultural, Food and Nutritional Science, University of Alberta, Edmonton, AB T6G2P5, Canada; [email protected] * Correspondence: [email protected]; Tel.: +1-850-412-5119; Fax: +1-850-561-2248 Abstract: The eastern oysters serve as important keystone species in the United States, especially in the Gulf of Mexico estuarine waters, and at the same time, provide unparalleled economic, ecological, environmental, and cultural services. One ecosystem service that has garnered recent attention is the ability of oysters to sequester impurities and nutrients, such as nitrogen (N), from the estuarine water that feeds them, via their exceptional filtration mechanism coupled with microbially-mediated denitrification processes. It is the oyster-associated microbiomes that essentially provide these myriads of ecological functions, yet not much is known on these microbiota at the genomic scale, especially from warm temperate and tropical water habitats. Among the suite of bacterial genera that appear to interplay with the oyster host species, pseudomonads deserve further assessment because Citation: Pathak, A.; Stothard, P.; of their immense metabolic and ecological potential. To obtain a comprehensive understanding on Chauhan, A. Comparative Genomic this aspect, we previously reported on the isolation and preliminary genomic characterization of Analysis of Three Pseudomonas Species three Pseudomonas species isolated from minced oyster tissue (P. -
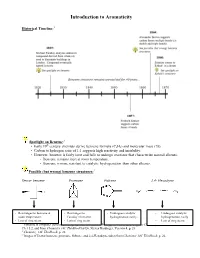
Introduction to Aromaticity
Introduction to Aromaticity Historical Timeline:1 Spotlight on Benzene:2 th • Early 19 century chemists derive benzene formula (C6H6) and molecular mass (78). • Carbon to hydrogen ratio of 1:1 suggests high reactivity and instability. • However, benzene is fairly inert and fails to undergo reactions that characterize normal alkenes. - Benzene remains inert at room temperature. - Benzene is more resistant to catalytic hydrogenation than other alkenes. Possible (but wrong) benzene structures:3 Dewar benzene Prismane Fulvene 2,4- Hexadiyne - Rearranges to benzene at - Rearranges to - Undergoes catalytic - Undergoes catalytic room temperature. Faraday’s benzene. hydrogenation easily. hydrogenation easily - Lots of ring strain. - Lots of ring strain. - Lots of ring strain. 1 Timeline is computer-generated, compiled with information from pg. 594 of Bruice, Organic Chemistry, 4th Edition, Ch. 15.2, and from Chemistry 14C Thinkbook by Dr. Steven Hardinger, Version 4, p. 26 2 Chemistry 14C Thinkbook, p. 26 3 Images of Dewar benzene, prismane, fulvene, and 2,4-Hexadiyne taken from Chemistry 14C Thinkbook, p. 26. Kekulé’s solution: - “snake bites its own tail” (4) Problems with Kekulé’s solution: • If Kekulé’s structure were to have two chloride substituents replacing two hydrogen atoms, there should be a pair of 1,2-dichlorobenzene isomers: one isomer with single bonds separating the Cl atoms, and another with double bonds separating the Cl atoms. • These isomers were never isolated or detected. • Rapid equilibrium proposed, where isomers interconvert so quickly that they cannot be isolated or detected. • Regardless, Kekulé’s structure has C=C’s and normal alkene reactions are still expected. - But the unusual stability of benzene still unexplained. -

On the Harmonic Oscillator Model of Electron Delocalization (HOMED) Index and Its Application to Heteroatomic Π-Electron Systems
Symmetry 2010, 2, 1485-1509; doi:10.3390/sym2031485 OPEN ACCESS symmetry ISSN 2073-8994 www.mdpi.com/journal/symmetry Article On the Harmonic Oscillator Model of Electron Delocalization (HOMED) Index and its Application to Heteroatomic π-Electron Systems Ewa D. Raczyñska 1, *, Małgorzata Hallman 1, Katarzyna Kolczyñska 2 and Tomasz M. Stêpniewski 2 1 Department of Chemistry, Warsaw University of Life Sciences (SGGW), ul. Nowoursynowska 159c, 02-776 Warszawa, Poland 2 Interdisciplinary Department of Biotechnology, Warsaw University of Life Sciences (SGGW), ul. Nowoursynowska 166, 02-776 Warszawa, Poland * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +48-22-59-37623; Fax: +49-22-59-37635. Received: 23 April 2010; in revised form: 19 May 2010 / Accepted: 7 July 2010 / Published: 12 July 2010 Abstract: The HOMA (Harmonic Oscillator Model of Aromaticity) index, reformulated in 1993, has been very often applied to describe π-electron delocalization for mono- and polycyclic π-electron systems. However, different measures of π-electron delocalization were employed for the CC, CX, and XY bonds, and this index seems to be inappropriate for compounds containing heteroatoms. In order to describe properly various resonance effects (σ-π hyperconjugation, n-π conjugation, π-π conjugation, and aromaticity) possible for heteroatomic π-electron systems, some modifications, based on the original HOMA idea, were proposed and tested for simple DFT structures containing C, N, and O atoms. An abbreviation HOMED was used for the modified index. Keywords: geometry-based index; π -electron delocalization; σ - π hyperconjugation; n-π conjugation; π-π conjugation; aromaticity; heteroatomic compounds; DFT 1. -

Aromaticity Sem- Ii
AROMATICITY SEM- II In 1931, German chemist and physicist Sir Erich Hückel proposed a theory to help determine if a planar ring molecule would have aromatic properties .This is a very popular and useful rule to identify aromaticity in monocyclic conjugated compound. According to which a planar monocyclic conjugated system having ( 4n +2) delocalised (where, n = 0, 1, 2, .....) electrons are known as aromatic compound . For example: Benzene, Naphthalene, Furan, Pyrrole etc. Criteria for Aromaticity 1) The molecule is cyclic (a ring of atoms) 2) The molecule is planar (all atoms in the molecule lie in the same plane) 3) The molecule is fully conjugated (p orbitals at every atom in the ring) 4) The molecule has 4n+2 π electrons (n=0 or any positive integer Why 4n+2π Electrons? According to Hückel's Molecular Orbital Theory, a compound is particularly stable if all of its bonding molecular orbitals are filled with paired electrons. - This is true of aromatic compounds, meaning they are quite stable. - With aromatic compounds, 2 electrons fill the lowest energy molecular orbital, and 4 electrons fill each subsequent energy level (the number of subsequent energy levels is denoted by n), leaving all bonding orbitals filled and no anti-bonding orbitals occupied. This gives a total of 4n+2π electrons. - As for example: Benzene has 6π electrons. Its first 2π electrons fill the lowest energy orbital, and it has 4π electrons remaining. These 4 fill in the orbitals of the succeeding energy level. The criteria for Antiaromaticity are as follows: 1) The molecule must be cyclic and completely conjugated 2) The molecule must be planar. -

Insights Into the Codon Usage Bias of 13 Severe Acute
bioRxiv preprint doi: https://doi.org/10.1101/2020.04.01.019463; this version posted April 4, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Insights into The Codon Usage Bias of 13 Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Isolates from Different Geo- locations Ali Mostafa Anwar *1, Saif M. Khodary 1 1 Department of Genetics, Faculty of Agriculture, Cairo University, Giza, 12613, Egypt *Correspondence: [email protected] Abstract Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of Coronavirus disease 2019 (COVID-19) which is an infectious disease that spread throughout the world and was declared as a pandemic by the World Health Organization (WHO). In the present study, we analyzed genome-wide codon usage patterns in 13 SARS-CoV-2 isolates from different geo-locations (countries) by utilizing different CUB measurements. Nucleotide and di-nucleotide compositions displayed bias toward A/U content in all codon positions and CpU-ended codons preference, respectively. Relative Synonymous Codon Usage (RSCU) analysis revealed 8 common putative preferred codons among all the 13 isolates. Interestingly, all of the latter codons are A/U-ended (U-ended: 7, A-ended: 1). Cluster analysis (based on RSCU values) was performed and showed comparable results to the phylogenetic analysis (based on their whole genome sequences) indicating that the CUB pattern may reflect the evolutionary relationship between the tested isolates. -

Codon Usage and Adenovirus Fitness: Implications for Vaccine Development
fmicb-12-633946 February 8, 2021 Time: 11:48 # 1 REVIEW published: 10 February 2021 doi: 10.3389/fmicb.2021.633946 Codon Usage and Adenovirus Fitness: Implications for Vaccine Development Judit Giménez-Roig1, Estela Núñez-Manchón1, Ramon Alemany2, Eneko Villanueva3 and Cristina Fillat1,4,5* 1 Institut d’Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS), Barcelona, Spain, 2 Procure Program, Institut Català d’Oncologia- Oncobell Program, IDIBELL, L’Hospitalet de Llobregat, Barcelona, Spain, 3 Cambridge Centre for Proteomics, Department of Biochemistry, University of Cambridge, Cambridge, United Kingdom, 4 Centro de Investigación Biomédica en Red de Enfermedades Raras (CIBERER), Barcelona, Spain, 5 Facultat de Medicina i Ciències de la Salut, Universitat de Barcelona (UB), Barcelona, Spain Vaccination is the most effective method to date to prevent viral diseases. It intends to mimic a naturally occurring infection while avoiding the disease, exposing our bodies to viral antigens to trigger an immune response that will protect us from future infections. Among different strategies for vaccine development, recombinant vaccines are one of the most efficient ones. Recombinant vaccines use safe viral vectors as vehicles and incorporate a transgenic antigen of the pathogen against which we intend to Edited by: Rosa Maria Pintó, generate an immune response. These vaccines can be based on replication-deficient University of Barcelona, Spain viruses or replication-competent viruses. While the most effective strategy involves Reviewed by: replication-competent viruses, they must be attenuated to prevent any health hazard Kai Li, Harbin Veterinary Research Institute, while guaranteeing a strong humoral and cellular immune response. Several attenuation Chinese Academy of Agricultural strategies for adenoviral-based vaccine development have been contemplated over Sciences, China time. -

Pseudomonas Chloritidismutans Sp. Nov., a Non- Denitrifying, Chlorate-Reducing Bacterium
International Journal of Systematic and Evolutionary Microbiology (2002), 52, 2183–2190 DOI: 10.1099/ijs.0.02102-0 Pseudomonas chloritidismutans sp. nov., a non- denitrifying, chlorate-reducing bacterium Laboratory of Microbiology, A. F. W. M. Wolterink, A. B. Jonker, S. W. M. Kengen and A. J. M. Stams Wageningen University, H. van Suchtelenweg 4, 6703 CT Wageningen, Author for correspondence: The Netherlands A. F. W. M. Wolterink. Tel: j31 317 484099. Fax: j31 317 483829. e-mail: Arthur.Wolterink!algemeen.micr.wag-ur.nl A Gram-negative, facultatively anaerobic, rod-shaped, dissimilatory chlorate- reducing bacterium, strain AW-1T, was isolated from biomass of an anaerobic chlorate-reducing bioreactor. Phylogenetic analysis of the 16S rDNA sequence showed 100% sequence similarity to Pseudomonas stutzeri DSM 50227 and 986% sequence similarity to the type strain of P. stutzeri (DSM 5190T). The species P. stutzeri possesses a high degree of genotypic and phenotypic heterogeneity. Therefore, eight genomic groups, termed genomovars, have been proposed based upon ∆Tm values, which were used to evaluate the quality of the pairing within heteroduplexes formed by DNA–DNA hybridization. In this study, DNA–DNA hybridization between strain AW-1T and P. stutzeri strains DSM 50227 and DSM 5190T revealed respectively 805 and 565% similarity. DNA–DNA hybridization between P. stutzeri strains DSM 50227 and DSM 5190T revealed 484% similarity. DNA–DNA hybridization indicated that strain AW-1T is not related at the species level to the type strain of P. stutzeri. However, strain AW-1T and P. stutzeri DSM 50227 are related at the species level. The physiological and biochemical properties of strain AW-1T and the two P. -

Aromaticity in Polyhydride Complexes of Ru, Ir, Os, and Pt† Cite This: DOI: 10.1039/C5cp04330a Elisa Jimenez-Izala and Anastassia N
PCCP View Article Online PAPER View Journal r-Aromaticity in polyhydride complexes of Ru, Ir, Os, and Pt† Cite this: DOI: 10.1039/c5cp04330a Elisa Jimenez-Izala and Anastassia N. Alexandrova*ab Transition-metal hydrides represent a unique class of compounds, which are essential for catalysis, organic i À i À synthesis, and hydrogen storage. In this work we study IrH5(PPh3)2,(RuH5(P Pr3)2) ,(OsH5(PPr3)2) ,and À OsH4(PPhMe2)3 polyhydride complexes, inspired by the recent discovery of the s-aromatic PtZnH5 cluster À anion. The distinctive feature of these molecules is that, like in the PtZnH5 cluster, the metal is five-fold Received 23rd July 2015, coordinated in-plane, and holds additional ligands attheaxialpositions.Thisworkshowsthattheunusual Accepted 17th September 2015 coordination in these compounds indeed can be explained by s-aromaticity in the pentagonal arrangement, DOI: 10.1039/c5cp04330a stabilized by the atomic orbitals on the metal. Based on this newly elucidated bonding principle, we additionally propose a new family of polyhydrides that display a uniquely high coordination. We also report the first www.rsc.org/pccp indications of how aromaticity may impact the reactivity of these molecules. 1 Introduction many catalytic cycles, where they have either been used as catalysts or invoked as key intermediates.15,16 Aromaticity in Aromaticity is a very important concept in the contemporary general is associated with high symmetry, stability, and specific chemistry: the particular electronic structure of aromatic reactivity. Therefore, the stability of transition-metal hydride systems, where electrons are delocalized, explains their large complexes might have an important repercussion in the kinetics energetic stabilization and low reactivity. -

Étude Des Communautés Microbiennes Rhizosphériques De Ligneux Indigènes De Sols Anthropogéniques, Issus D’Effluents Industriels Cyril Zappelini
Étude des communautés microbiennes rhizosphériques de ligneux indigènes de sols anthropogéniques, issus d’effluents industriels Cyril Zappelini To cite this version: Cyril Zappelini. Étude des communautés microbiennes rhizosphériques de ligneux indigènes de sols anthropogéniques, issus d’effluents industriels. Sciences agricoles. Université Bourgogne Franche- Comté, 2018. Français. NNT : 2018UBFCD057. tel-01902775 HAL Id: tel-01902775 https://tel.archives-ouvertes.fr/tel-01902775 Submitted on 23 Oct 2018 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. UNIVERSITÉ DE BOURGOGNE FRANCHE-COMTÉ École doctorale Environnement-Santé Laboratoire Chrono-Environnement (UMR UFC/CNRS 6249) THÈSE Présentée en vue de l’obtention du titre de Docteur de l’Université Bourgogne Franche-Comté Spécialité « Sciences de la Vie et de l’Environnement » ÉTUDE DES COMMUNAUTES MICROBIENNES RHIZOSPHERIQUES DE LIGNEUX INDIGENES DE SOLS ANTHROPOGENIQUES, ISSUS D’EFFLUENTS INDUSTRIELS Présentée et soutenue publiquement par Cyril ZAPPELINI Le 3 juillet 2018, devant le jury composé de : Membres du jury : Vera SLAVEYKOVA (Professeure, Univ. de Genève) Rapporteure Bertrand AIGLE (Professeur, Univ. de Lorraine) Rapporteur & président du jury Céline ROOSE-AMSALEG (IGR, Univ. de Rennes) Examinatrice Karine JEZEQUEL (Maître de conférences, Univ. de Haute Alsace) Examinatrice Nicolas CAPELLI (Maître de conférences HDR, UBFC) Encadrant Christophe GUYEUX (Professeur, UBFC) Co-directeur de thèse Michel CHALOT (Professeur, UBFC) Directeur de thèse « En vérité, le chemin importe peu, la volonté d'arriver suffit à tout. -

Codon Clusters with Biased Synonymous Codon Usage Represent
bioRxiv preprint doi: https://doi.org/10.1101/530345; this version posted January 25, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Codon clusters with biased synonymous codon usage represent 2 hidden functional domains in protein-coding DNA sequences 3 4 Authors 5 Zhen Peng1*, Yehuda Ben-Shahar1 6 7 Affiliations 8 1Department of Biology, Washington University in St. Louis, MO 63130, USA. 9 *Correspondence to: Zhen Peng ([email protected]). 10 11 Outline 12 1. Abstract 13 2. Introduction 14 3. Results 15 3.1. Identifying putatively functional codon clusters (PFCCs) 16 3.2. Codon usage patterns of PFCCs are diverse 17 3.3. PFCC distribution is not restricted to specific regions of protein-coding sequences 18 3.4. Specific protein functional classes are overrepresented in genes carrying PFCCs while most PFCCs 19 are not associated with known protein domains 20 3.5. Voltage-gated sodium channels include a conserved rare-codon cluster associated with the 21 inactivation gate 22 4. Discussion 23 5. Materials and Methods 24 5.1. Reference genome and data pre-procession Page 1 of 28 bioRxiv preprint doi: https://doi.org/10.1101/530345; this version posted January 25, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 25 5.2. Identifying PFCCs 26 5.3. Calculating TCAI 27 5.4. K-mean clustering of PFCCs 28 5.5. -

Pseudomonas Stutzeri Biology Of
Biology of Pseudomonas stutzeri Jorge Lalucat, Antoni Bennasar, Rafael Bosch, Elena García-Valdés and Norberto J. Palleroni Microbiol. Mol. Biol. Rev. 2006, 70(2):510. DOI: 10.1128/MMBR.00047-05. Downloaded from Updated information and services can be found at: http://mmbr.asm.org/content/70/2/510 These include: http://mmbr.asm.org/ REFERENCES This article cites 395 articles, 145 of which can be accessed free at: http://mmbr.asm.org/content/70/2/510#ref-list-1 CONTENT ALERTS Receive: RSS Feeds, eTOCs, free email alerts (when new articles cite this article), more» on January 28, 2014 by Red de Bibliotecas del CSIC Information about commercial reprint orders: http://journals.asm.org/site/misc/reprints.xhtml To subscribe to to another ASM Journal go to: http://journals.asm.org/site/subscriptions/ MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS, June 2006, p. 510–547 Vol. 70, No. 2 1092-2172/06/$08.00ϩ0 doi:10.1128/MMBR.00047-05 Copyright © 2006, American Society for Microbiology. All Rights Reserved. Biology of Pseudomonas stutzeri Jorge Lalucat,1,2* Antoni Bennasar,1 Rafael Bosch,1 Elena Garcı´a-Valde´s,1,2 and Norberto J. Palleroni3 Departament de Biologia, Microbiologia, Universitat de les Illes Balears, Campus UIB, 07122 Palma de Mallorca, Spain1; Institut Mediterrani d’Estudis Avanc¸ats (CSIC-UIB), Campus UIB, 07122 Palma de Mallorca, Spain2; and Department of Biochemistry and Microbiology, Rutgers University, Cook Campus, 3 New Brunswick, New Jersey 08901-8520 Downloaded from INTRODUCTION .......................................................................................................................................................511