Molecular Diagnosis of Mitochondrial Disorders
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Molecular Diagnostic Requisition
BAYLOR MIRACA GENETICS LABORATORIES SHIP TO: Baylor Miraca Genetics Laboratories 2450 Holcombe, Grand Blvd. -Receiving Dock PHONE: 800-411-GENE | FAX: 713-798-2787 | www.bmgl.com Houston, TX 77021-2024 Phone: 713-798-6555 MOLECULAR DIAGNOSTIC REQUISITION PATIENT INFORMATION SAMPLE INFORMATION NAME: DATE OF COLLECTION: / / LAST NAME FIRST NAME MI MM DD YY HOSPITAL#: ACCESSION#: DATE OF BIRTH: / / GENDER (Please select one): FEMALE MALE MM DD YY SAMPLE TYPE (Please select one): ETHNIC BACKGROUND (Select all that apply): UNKNOWN BLOOD AFRICAN AMERICAN CORD BLOOD ASIAN SKELETAL MUSCLE ASHKENAZIC JEWISH MUSCLE EUROPEAN CAUCASIAN -OR- DNA (Specify Source): HISPANIC NATIVE AMERICAN INDIAN PLACE PATIENT STICKER HERE OTHER JEWISH OTHER (Specify): OTHER (Please specify): REPORTING INFORMATION ADDITIONAL PROFESSIONAL REPORT RECIPIENTS PHYSICIAN: NAME: INSTITUTION: PHONE: FAX: PHONE: FAX: NAME: EMAIL (INTERNATIONAL CLIENT REQUIREMENT): PHONE: FAX: INDICATION FOR STUDY SYMPTOMATIC (Summarize below.): *FAMILIAL MUTATION/VARIANT ANALYSIS: COMPLETE ALL FIELDS BELOW AND ATTACH THE PROBAND'S REPORT. GENE NAME: ASYMPTOMATIC/POSITIVE FAMILY HISTORY: (ATTACH FAMILY HISTORY) MUTATION/UNCLASSIFIED VARIANT: RELATIONSHIP TO PROBAND: THIS INDIVIDUAL IS CURRENTLY: SYMPTOMATIC ASYMPTOMATIC *If family mutation is known, complete the FAMILIAL MUTATION/ VARIANT ANALYSIS section. NAME OF PROBAND: ASYMPTOMATIC/POPULATION SCREENING RELATIONSHIP TO PROBAND: OTHER (Specify clinical findings below): BMGL LAB#: A COPY OF ORIGINAL RESULTS ATTACHED IF PROBAND TESTING WAS PERFORMED AT ANOTHER LAB, CALL TO DISCUSS PRIOR TO SENDING SAMPLE. A POSITIVE CONTROL MAY BE REQUIRED IN SOME CASES. REQUIRED: NEW YORK STATE PHYSICIAN SIGNATURE OF CONSENT I certify that the patient specified above and/or their legal guardian has been informed of the benefits, risks, and limitations of the laboratory test(s) requested. -

Yeast and Rat Coq3 and Escherichia Coli Ubig Polypeptides Catalyze Both O-Methyltransferase Steps in Coenzyme Q Biosynthesis*
THE JOURNAL OF BIOLOGICAL CHEMISTRY Vol. 274, No. 31, Issue of July 30, pp. 21665–21672, 1999 © 1999 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in U.S.A. Yeast and Rat Coq3 and Escherichia coli UbiG Polypeptides Catalyze Both O-Methyltransferase Steps in Coenzyme Q Biosynthesis* (Received for publication, April 27, 1999) Wayne W. Poon, Robert J. Barkovich, Adam Y. Hsu, Adam Frankel, Peter T. Lee, Jennifer N. Shepherd‡, David C. Myles, and Catherine F. Clarke§ From the Department of Chemistry and Biochemistry and the Molecular Biology Institute, University of California, Los Angeles, California 90095 Ubiquinone (coenzyme Q or Q) is a lipid that functions the isoprenoid tail functions to anchor Q in the membrane. In in the electron transport chain in the inner mitochon- eukaryotes, Q functions to shuttle electrons from either Com- drial membrane of eukaryotes and the plasma mem- plex I or Complex II to Complex III/bc1 complex. The transfer of brane of prokaryotes. Q-deficient mutants of Saccharo- electrons from Q to the bc1 complex is coupled to proton-trans- myces cerevisiae harbor defects in one of eight COQ location via the Q cycle mechanism that was first proposed by genes (coq1–coq8) and are unable to grow on nonfer- Mitchell (2). A number of studies support such a mechanism mentable carbon sources. The biosynthesis of Q involves (for a review, see Ref. 1) including the recently determined two separate O-methylation steps. In yeast, the first O- complete structure of the bc1 complex (3). methylation utilizes 3,4-dihydroxy-5-hexaprenylbenzoic The redox properties of Q also allow it to function as a lipid acid as a substrate and is thought to be catalyzed by soluble antioxidant. -

Metabolic Targets of Coenzyme Q10 in Mitochondria
antioxidants Review Metabolic Targets of Coenzyme Q10 in Mitochondria Agustín Hidalgo-Gutiérrez 1,2,*, Pilar González-García 1,2, María Elena Díaz-Casado 1,2, Eliana Barriocanal-Casado 1,2, Sergio López-Herrador 1,2, Catarina M. Quinzii 3 and Luis C. López 1,2,* 1 Departamento de Fisiología, Facultad de Medicina, Universidad de Granada, 18016 Granada, Spain; [email protected] (P.G.-G.); [email protected] (M.E.D.-C.); [email protected] (E.B.-C.); [email protected] (S.L.-H.) 2 Centro de Investigación Biomédica, Instituto de Biotecnología, Universidad de Granada, 18016 Granada, Spain 3 Department of Neurology, Columbia University Medical Center, New York, NY 10032, USA; [email protected] * Correspondence: [email protected] (A.H.-G.); [email protected] (L.C.L.); Tel.: +34-958-241-000 (ext. 20197) (L.C.L.) Abstract: Coenzyme Q10 (CoQ10) is classically viewed as an important endogenous antioxidant and key component of the mitochondrial respiratory chain. For this second function, CoQ molecules seem to be dynamically segmented in a pool attached and engulfed by the super-complexes I + III, and a free pool available for complex II or any other mitochondrial enzyme that uses CoQ as a cofactor. This CoQ-free pool is, therefore, used by enzymes that link the mitochondrial respiratory chain to other pathways, such as the pyrimidine de novo biosynthesis, fatty acid β-oxidation and amino acid catabolism, glycine metabolism, proline, glyoxylate and arginine metabolism, and sulfide oxidation Citation: Hidalgo-Gutiérrez, A.; metabolism. Some of these mitochondrial pathways are also connected to metabolic pathways González-García, P.; Díaz-Casado, in other compartments of the cell and, consequently, CoQ could indirectly modulate metabolic M.E.; Barriocanal-Casado, E.; López-Herrador, S.; Quinzii, C.M.; pathways located outside the mitochondria. -

Clinical Laboratory Services
The Commonwealth of Massachusetts Executive Office of Health and Human Services Office of Medicaid One Ashburton Place, Room 1109 Boston, Massachusetts 02108 Administrative Bulletin 15-03 DANIEL TSAI CHARLES D. BAKER Governor 101 CMR 320.00: Clinical Laboratory Assistant Secretary for MassHealth KARYN E. POLITO Services Lieutenant Governor Tel: (617) 573-1600 Effective January 1, 2015 MARYLOU SUDDERS Fax: (617) 573-1891 Secretary www.mass.gov/eohhs Procedure Code Update Under the authority of regulation 101 CMR 320.01(3), the Executive Office of Health and Human Services is adding new procedure codes and is deleting outdated codes. The rates for code additions are priced at 74.67% of the prevailing Medicare fee if available. The changes, effective January 1, 2015, are as follows. Code Change Rate Code Description (if applicable) 80100 Deletion 80101 Deletion 80103 Deletion 80104 Deletion 80163 Addition $13.49 Digoxin; free 80165 Addition $13.77 Valproic acid (dipropylacetic acid); total 80440 Deletion 81246 Addition I.C. FLT3 (fms-related tyrosine kinase 3) (e.g., acute myeloid leukemia), gene analysis; internal tandem duplication (ITD) variants (IE, exons 14,15) 81288 Addition I.C. MLH1 (mutL homolog 1, colon cancer, nonpolyposis type 2) (e.g., hereditary non-polyposis colorectal cancer, Lynch syndrome) gene analysis; promoter methylation analysis 81313 Addition I.C. PCA3/KLK3 (prostate cancer antigen 3 [non-protein coding]/kallikrien- related peptidase 3 [prostate specific antigen]) ratio (e.g., prostate cancer) 81410 Addition I.C. Aortic dysfunction or dilation (e.g., Marfan syndrome, Loeys Dietz syndrome, Ehler Danlos syndrome type IV, arterial tortuosity syndrome); genomic sequence analysis panel, must include sequencing of at least 9 genes, including FBN1, TGFBR1, TGFBR2, COL3A1, MYH11, ACTA2, SLC2A10, SMAD3, and MYLK 81411 Addition I.C. -

Human Mitochondrial Pathologies of the Respiratory Chain and ATP Synthase: Contributions from Studies of Saccharomyces Cerevisiae
life Review Human Mitochondrial Pathologies of the Respiratory Chain and ATP Synthase: Contributions from Studies of Saccharomyces cerevisiae Leticia V. R. Franco 1,2,* , Luca Bremner 1 and Mario H. Barros 2 1 Department of Biological Sciences, Columbia University, New York, NY 10027, USA; [email protected] 2 Department of Microbiology,Institute of Biomedical Sciences, Universidade de Sao Paulo, Sao Paulo 05508-900, Brazil; [email protected] * Correspondence: [email protected] Received: 27 October 2020; Accepted: 19 November 2020; Published: 23 November 2020 Abstract: The ease with which the unicellular yeast Saccharomyces cerevisiae can be manipulated genetically and biochemically has established this organism as a good model for the study of human mitochondrial diseases. The combined use of biochemical and molecular genetic tools has been instrumental in elucidating the functions of numerous yeast nuclear gene products with human homologs that affect a large number of metabolic and biological processes, including those housed in mitochondria. These include structural and catalytic subunits of enzymes and protein factors that impinge on the biogenesis of the respiratory chain. This article will review what is currently known about the genetics and clinical phenotypes of mitochondrial diseases of the respiratory chain and ATP synthase, with special emphasis on the contribution of information gained from pet mutants with mutations in nuclear genes that impair mitochondrial respiration. Our intent is to provide the yeast mitochondrial specialist with basic knowledge of human mitochondrial pathologies and the human specialist with information on how genes that directly and indirectly affect respiration were identified and characterized in yeast. Keywords: mitochondrial diseases; respiratory chain; yeast; Saccharomyces cerevisiae; pet mutants 1. -

COQ2 Gene Coenzyme Q2, Polyprenyltransferase
COQ2 gene coenzyme Q2, polyprenyltransferase Normal Function The COQ2 gene provides instructions for making an enzyme that carries out one step in the production of a molecule called coenzyme Q10, which has several critical functions in cells throughout the body. In cell structures called mitochondria, coenzyme Q10 plays an essential role in a process called oxidative phosphorylation, which converts the energy from food into a form cells can use. Coenzyme Q10 is also involved in producing pyrimidines, which are building blocks of DNA, its chemical cousin RNA, and molecules such as ATP and GTP that serve as energy sources in the cell. In cell membranes, coenzyme Q10 acts as an antioxidant, protecting cells from damage caused by unstable oxygen-containing molecules (free radicals), which are byproducts of energy production. Health Conditions Related to Genetic Changes Multiple system atrophy Several variations in the COQ2 gene have been suggested to increase the risk of multiple system atrophy, a progressive brain disorder that affects movement and balance and disrupts the function of the autonomic nervous system. The autonomic nervous system controls body functions that are mostly involuntary, such as regulation of blood pressure. The identified variations alter single protein building blocks (amino acids) in the COQ2 enzyme. Most of the variations are very rare, but a genetic change that replaces the amino acid valine with the amino acid alanine at position 393 (written as Val393Ala or V393A) is relatively common. Studies suggest that these variations, including V393A, are associated with an increased risk of developing multiple system atrophy in the Japanese population. However, studies have not found a correlation between COQ2 gene variations and multiple system atrophy in other populations, including Koreans, Europeans, and North Americans. -

Mitochondrial Diseases: Expanding the Diagnosis in the Era of Genetic Testing
Saneto. J Transl Genet Genom 2020;4:384-428 Journal of Translational DOI: 10.20517/jtgg.2020.40 Genetics and Genomics Review Open Access Mitochondrial diseases: expanding the diagnosis in the era of genetic testing Russell P. Saneto1,2 1Center for Integrative Brain Research, Neuroscience Institute, Seattle, WA 98101, USA. 2Department of Neurology/Division of Pediatric Neurology, Seattle Children’s Hospital/University of Washington, Seattle, WA 98105, USA. Correspondence to: Dr. Russell P. Saneto, Department of Neurology/Division of Pediatric Neurology, Seattle Children’s Hospital/ University of Washington, 4800 Sand Point Way NE, Seattle, WA 98105, USA. E-mail: [email protected] How to cite this article: Saneto RP. Mitochondrial diseases: expanding the diagnosis in the era of genetic testing. J Transl Genet Genom 2020;4:348-428. http://dx.doi.org/10.20517/jtgg.2020.40 Received: 29 Jun 2020 First Decision: 27 Jul 2020 Revised: 15 Aug 2020 Accepted: 21 Aug 2020 Available online: 29 Sep 2020 Academic Editor: Andrea L. Gropman Copy Editor: Cai-Hong Wang Production Editor: Jing Yu Abstract Mitochondrial diseases are clinically and genetically heterogeneous. These diseases were initially described a little over three decades ago. Limited diagnostic tools created disease descriptions based on clinical, biochemical analytes, neuroimaging, and muscle biopsy findings. This diagnostic mechanism continued to evolve detection of inherited oxidative phosphorylation disorders and expanded discovery of mitochondrial physiology over the next two decades. Limited genetic testing hampered the definitive diagnostic identification and breadth of diseases. Over the last decade, the development and incorporation of massive parallel sequencing has identified approximately 300 genes involved in mitochondrial disease. -
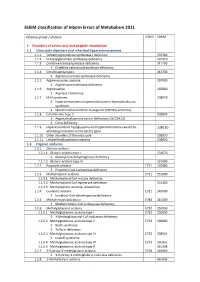
SSIEM Classification of Inborn Errors of Metabolism 2011
SSIEM classification of Inborn Errors of Metabolism 2011 Disease group / disease ICD10 OMIM 1. Disorders of amino acid and peptide metabolism 1.1. Urea cycle disorders and inherited hyperammonaemias 1.1.1. Carbamoylphosphate synthetase I deficiency 237300 1.1.2. N-Acetylglutamate synthetase deficiency 237310 1.1.3. Ornithine transcarbamylase deficiency 311250 S Ornithine carbamoyltransferase deficiency 1.1.4. Citrullinaemia type1 215700 S Argininosuccinate synthetase deficiency 1.1.5. Argininosuccinic aciduria 207900 S Argininosuccinate lyase deficiency 1.1.6. Argininaemia 207800 S Arginase I deficiency 1.1.7. HHH syndrome 238970 S Hyperammonaemia-hyperornithinaemia-homocitrullinuria syndrome S Mitochondrial ornithine transporter (ORNT1) deficiency 1.1.8. Citrullinemia Type 2 603859 S Aspartate glutamate carrier deficiency ( SLC25A13) S Citrin deficiency 1.1.9. Hyperinsulinemic hypoglycemia and hyperammonemia caused by 138130 activating mutations in the GLUD1 gene 1.1.10. Other disorders of the urea cycle 238970 1.1.11. Unspecified hyperammonaemia 238970 1.2. Organic acidurias 1.2.1. Glutaric aciduria 1.2.1.1. Glutaric aciduria type I 231670 S Glutaryl-CoA dehydrogenase deficiency 1.2.1.2. Glutaric aciduria type III 231690 1.2.2. Propionic aciduria E711 232000 S Propionyl-CoA-Carboxylase deficiency 1.2.3. Methylmalonic aciduria E711 251000 1.2.3.1. Methylmalonyl-CoA mutase deficiency 1.2.3.2. Methylmalonyl-CoA epimerase deficiency 251120 1.2.3.3. Methylmalonic aciduria, unspecified 1.2.4. Isovaleric aciduria E711 243500 S Isovaleryl-CoA dehydrogenase deficiency 1.2.5. Methylcrotonylglycinuria E744 210200 S Methylcrotonyl-CoA carboxylase deficiency 1.2.6. Methylglutaconic aciduria E712 250950 1.2.6.1. Methylglutaconic aciduria type I E712 250950 S 3-Methylglutaconyl-CoA hydratase deficiency 1.2.6.2. -

Carrier Screening Panel
Advancing Non-Invasive Healthcare Cell3TM Target: Carrier Screening Panel Detects 448 childhood recessive disorders DISORDER GENE DISEASE TYPE 2-Methylbutryl-CoA dehydrogenase deficiency ACADSB Metabolic 3-Hydroxy-3-methylglutaryl- CoA lyase deficiency HMGCL Metabolic 3-Hydroxyacyl- CoA dehydrogenase deficiency HADH Metabolic 3-Methylcrotonyl- CoA carboxylase 2 deficiency MCCC2 Metabolic 3-Methylglutaconic aciduria, i AUH Metabolic 3-Methylglutaconic aciduria, iii OPA3 Metabolic 3-Methylglutaconic aciduria, v DNAJC19 Metabolic α -Thalassemia/mental retardation syndrome, nondeletion, XLR ATRX ATRX Hematologic ABCD syndrome EDNRB Cutaneous Abetalipoproteinemia; ABL MTTP Metabolic Achalasia-Addisonianism-Alacrima syndrome; AAA AAAS Endocrine Achondrogenesis, Ib; ACG1b SLC26A2 Skeletal Achromatopsia 3; ACHM3 CNGB3 Ocular Acrocallosal syndrome; ACLS GLI3 Developmental Acyl-CoA dehydrogenase family, member 9, deficiency of ACAD9 Metabolic Acyl-CoA dehydrogenase, long-chain, deficiency of ACADL Metabolic Acyl-CoA dehydrogenase, medium-chain, deficiency of ACADM Metabolic Acyl-CoA dehydrogenase, short-chain, deficiency of ACADS Metabolic Acyl-CoA dehydrogenase, very long-chain, deficiency of ACADVL Metabolic Adrenal hyperplasia, congenital, due to 21-hydroxylase deficiency CYP21A2 Endocrine Adrenoleukodystrophy; ALD ABCD1 Neurological Afibrinogenemia, congenital FGA Hematologic Afibrinogenemia, congenital FGB Hematologic Afibrinogenemia, congenital FGG Hematologic Agammaglobulinemia, XLR, XLA BTK Immunodeficiency Agenesis of the corpus callosum -

Mutations in Ubiquinone Deficiency and Oxidative Phosphorylation Disorders
Prenyldiphosphate synthase, subunit 1 (PDSS1) and OH- benzoate polyprenyltransferase (COQ2) mutations in ubiquinone deficiency and oxidative phosphorylation disorders Julie Mollet, … , Arnold Munnich, Agnès Rötig J Clin Invest. 2007;117(3):765-772. https://doi.org/10.1172/JCI29089. Research Article Coenzyme Q10 (CoQ10) plays a pivotal role in oxidative phosphorylation (OXPHOS), as it distributes electrons among the various dehydrogenases and the cytochrome segments of the respiratory chain. We have identified 2 novel inborn errors of CoQ10 biosynthesis in 2 distinct families. In both cases, enzymologic studies showed that quinone-dependent OXPHOS activities were in the range of the lowest control values, while OXPHOS enzyme activities were normal. CoQ10 deficiency was confirmed by restoration of normal OXPHOS activities after addition of quinone. A genome-wide search for homozygosity in family 1 identified a region of chromosome 10 encompassing the gene prenyldiphosphate synthase, subunit 1 (PDSS1), which encodes the human ortholog of the yeastC OQ1 gene, a key enzyme of CoQ10 synthesis. Sequencing of PDSS1 identified a homozygous nucleotide substitution modifying a conserved amino acid of the protein (D308E). In the second family, direct sequencing of OH-benzoate polyprenyltransferase (COQ2), the human ortholog of the yeast COQ2 gene, identified a single base pair frameshift deletion resulting in a premature stop codon (c.1198delT, N401fsX415). Transformation of yeast Δcoq1 and Δcoq2 strains by mutant yeast COQ1 and mutant human COQ2 -

Respiratory Chain Activity and Protein Expression in Skeletal Muscles from High and Low Feed Efficient Swine Within the Same Genetic Line
University of Arkansas, Fayetteville ScholarWorks@UARK Theses and Dissertations 7-2020 Respiratory Chain Activity and Protein Expression in Skeletal Muscles from High and Low Feed Efficient Swine within the Same Genetic Line Earsie L. Alcorn de Feliz University of Arkansas, Fayetteville Follow this and additional works at: https://scholarworks.uark.edu/etd Part of the Animal Experimentation and Research Commons, Cellular and Molecular Physiology Commons, Food Science Commons, and the Meat Science Commons Citation Alcorn de Feliz, E. L. (2020). Respiratory Chain Activity and Protein Expression in Skeletal Muscles from High and Low Feed Efficient Swine within the Same Genetic Line. Theses and Dissertations Retrieved from https://scholarworks.uark.edu/etd/3802 This Dissertation is brought to you for free and open access by ScholarWorks@UARK. It has been accepted for inclusion in Theses and Dissertations by an authorized administrator of ScholarWorks@UARK. For more information, please contact [email protected]. Respiratory Chain Activity and Protein Expression in Skeletal Muscles from High and Low Feed Efficient Swine within the Same Genetic Line A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Food Science by Earsie L. Alcorn de Feliz Union University Bachelor of Science in Biology, 1975 University of Arkansas Master of Science in Natural Science, 1977 July 2020 University of Arkansas This dissertation is approved for recommendation to the graduate council. Charles V. Maxwell, Ph.D. Dissertation Director Navam Hettiarachchy, Ph. D. Gisela F. Erf, Ph.D. Committee Member Committee Member Zelpha B. Johnson, Ph.D. Luke Howard, Ph.D. Committee Member Committee Member ABSTRACT Current as well as past studies have provided evidence for a connection between macroeconomic/agricultural and cellular/biochemical basis of feed efficiency (FE; gain:feed) in farm animal production. -

Combined Defects in Oxidative Phosphorylation and Fatty Acid Β-Oxidation in Mitochondrial Disease
Biosci. Rep. (2016) / 36 / art:e00313 / doi 10.1042/BSR20150295 Combined defects in oxidative phosphorylation and fatty acid β-oxidation in mitochondrial disease Abena Nsiah-Sefaa*† and Matthew McKenzie*†1 *Centre for Genetic Diseases, Hudson Institute of Medical Research, Clayton, Victoria 3168, Australia †The Department of Molecular and Translational Science, Monash University, Clayton, Victoria 3168, Australia Synopsis Mitochondria provide the main source of energy to eukaryotic cells, oxidizing fats and sugars to generate ATP. Mitochondrial fatty acid β-oxidation (FAO) and oxidative phosphorylation (OXPHOS) are two metabolic pathways which are central to this process. Defects in these pathways can result in diseases of the brain, skeletal muscle, heart and liver, affecting approximately 1 in 5000 live births. There are no effective therapies for these disorders, with quality of life severely reduced for most patients. The pathology underlying many aspects of these diseases is not well understood; for example, it is not clear why some patients with primary FAO deficiencies exhibit secondary OXPHOS defects. However, recent findings suggest that physical interactions exist between FAO and OXPHOS proteins, and that these interactions are critical for both FAO and OXPHOS function. Here, we review our current understanding of the interactions between FAO and OXPHOS proteins and how defects in these two metabolic pathways contribute to mitochondrial disease pathogenesis. Key words: disease, mitochondria, protein complex assembly, protein interactions, supercomplex. Cite this article as: Bioscience Reports (2016) 36, e00313, doi:10.1042/BSR20150295 MITOCHONDRIAL METABOLISM the absence of oxygen in the cytoplasm (glycolysis), or by the metabolism of fats, sugars and proteins in the mitochon- dria in the presence of oxygen.