Comparative Transcriptome Analysis of Alpinia Oxyphylla Miq. Reveals
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
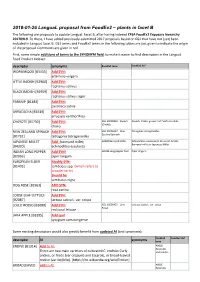
2018-01-26 Langual Proposal from Foodex2 – Plants in Facet B
2018-01-26 LanguaL proposal from FoodEx2 – plants in facet B The following are proposals to update LanguaL Facet B, after having indexed EFSA FoodEx2 Exposure hierarchy 20170919. To these, I have added previously-submitted 2017 proposals based on GS1 that have not (yet) been included in LanguaL facet B. GS1 terms and FoodEx2 terms in the following tables are just given to indicate the origin of the proposal. Comments are given in red. First, some simple additions of terms to the SYNONYM field, to make it easier to find descriptors in the LanguaL Food Product Indexer: descriptor synonyms FoodEx2 term FoodEx2 def WORMWOOD [B3433] Add SYN: artemisia vulgaris LITTLE RADISH [B2960] Add SYN: raphanus sativus BLACK RADISH [B2959] Add SYN: raphanus sativus niger PARSNIP [B1483] Add SYN: pastinaca sativa ARRACACHA [B3439] Add SYN: arracacia xanthorrhiza CHAYOTE [B1730] Add SYN: GS1 10006356 - Squash Squash, Choko, grown from Sechium edule (Choko) choko NEW ZEALAND SPINACH Add SYN: GS1 10006427 - New- Tetragonia tetragonoides Zealand Spinach [B1732] tetragonia tetragonoides JAPANESE MILLET Add : barnyard millet; A000Z Barnyard millet Echinochloa esculenta (A. Braun) H. Scholz, Barnyard millet or Japanese Millet. [B4320] echinochloa esculenta INDIAN LONG PEPPER Add SYN! A019B Long pepper fruit Piper longum [B2956] piper longum EUROPEAN ELDER Modify SYN: [B1403] sambucus spp. (which refers to broader term) Should be sambucus nigra DOG ROSE [B2961] ADD SYN: rosa canina LOOSE LEAF LETTUCE Add SYN: [B2087] lactusa sativa L. var. crispa LOLLO ROSSO [B2088] Add SYN: GS1 10006425 - Lollo Lactuca sativa L. var. crispa Rosso red coral lettuce JAVA APPLE [B3395] Add syn! syzygium samarangense Some existing descriptors would also greatly benefit from updated AI (and synonyms): FoodEx2 FoodEx2 def descriptor AI synonyms term ENDIVE [B1314] Add to AI: A00LD Escaroles There are two main varieties of cultivated C. -

Herbs, Spices and Essential Oils
Printed in Austria V.05-91153—March 2006—300 Herbs, spices and essential oils Post-harvest operations in developing countries UNITED NATIONS INDUSTRIAL DEVELOPMENT ORGANIZATION Vienna International Centre, P.O. Box 300, 1400 Vienna, Austria Telephone: (+43-1) 26026-0, Fax: (+43-1) 26926-69 UNITED NATIONS FOOD AND AGRICULTURE E-mail: [email protected], Internet: http://www.unido.org INDUSTRIAL DEVELOPMENT ORGANIZATION OF THE ORGANIZATION UNITED NATIONS © UNIDO and FAO 2005 — First published 2005 All rights reserved. Reproduction and dissemination of material in this information product for educational or other non-commercial purposes are authorized without any prior written permission from the copyright holders provided the source is fully acknowledged. Reproduction of material in this information product for resale or other commercial purposes is prohibited without written permission of the copyright holders. Applications for such permission should be addressed to: - the Director, Agro-Industries and Sectoral Support Branch, UNIDO, Vienna International Centre, P.O. Box 300, 1400 Vienna, Austria or by e-mail to [email protected] - the Chief, Publishing Management Service, Information Division, FAO, Viale delle Terme di Caracalla, 00100 Rome, Italy or by e-mail to [email protected] The designations employed and the presentation of material in this information product do not imply the expression of any opinion whatsoever on the part of the United Nations Industrial Development Organization or of the Food and Agriculture Organization of the United Nations concerning the legal or development status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. -

Antioxidant and Antimicrobial Activity of Zingiberaceae Plants in Taiwan
Plant Foods Hum Nutr (2008) 63:15–20 DOI 10.1007/s11130-007-0063-7 ORIGINAL PAPER Antioxidant and Antimicrobial Activity of Zingiberaceae Plants in Taiwan I-Nan Chen & Chen-Chin Chang & Chang-Chai Ng & Chung-Yi Wang & Yuan-Tay Shyu & Tsu-Liang Chang Published online: 20 December 2007 # Springer Science + Business Media, LLC 2007 Abstract The rhizomes of the Zingiberaceae family are a Keywords Antimicrobial . Antioxidant . Ginger . vegetable widely used in many Asian countries, and their Reducing power . Zingiberaceae medicinal functions have been broadly discussed and accepted in many traditional recipes. In this study, 18 species of five genus of Zingiberaceae plants from Taiwan area were Introduction collected and analyzed for their functional properties. Methanolic extracts of the plants were analyzed for their The Zingiberaceous plants are characterized by their total phenol compounds, α,α-diphenyl-β-picrylhydrazyl tuberous or non-tuberous rhizomes, which have strong (DPPH) scavenging activity, and reducing power. Antimi- aromatic and medicinal properties. It is commonly known crobial activity of these samples was also determined. The as ginger, and exists in about 50 genera and 1,300 species results showed that the total phenol compounds of the Alpinia worldwide, distributed mainly in South and Southeast Asia genus averaged 17, 30 mg/g for Curcumas, and the highest, [1]. Turmeric of Zingiberaceous plants in powder form is 36.5 mg/g for Vanoverberghia sasakiana. Antioxidant widely applied as a food additive in many Asian countries. performances were best observed in Vanoverberghia and Medicinal functions for treatment of diseases such as Hedychium, both 89%, and DPPH scavenging activity diarrhea, coryza, dermatosis disorders and rheumatism are followed similar trends. -

The Inhibitory Effect of Some Iranian Plants Extracts on the Alpha Glucosidase
Iranian Journal of Basic Medical Sciences Vol. 11, No. 1, Spring 2008, 1 - 9 Received: Mar 2, 2008; Accepted: May 17, 2008 The Inhibitory Effect of Some Iranian Plants Extracts on the Alpha Glucosidase *1Ahmad Gholamhoseinian, 1Hossein Fallah, 2Fariba Sharifi-far, 3Mansour Mirtajaddini Abstract Objective Diabetes mellitus is manifested by hyperglycaemia. Different treatments such as diets and drugs are recommended for diabetes control. For various reasons in recent years traditional plant (herbal) therapies as prescribed by indigenous systems of medicine with different mechanisms have commonly been used. The digestive enzymes such as alpha glucosidase are among these herbal remedies. Materials and Methods One hundred species of plants were collected or purchased from the Medicinal Herbal Markets and botanically identified. Methanolic and aqueous extracts were prepared by the maceration method. The enzymatic activities of alpha glucosidase were determined colorimetrically by monitoring the release of p-nitrophenol from the appropriate p-nitrophenol glycoside substrate, after 30 mins incubation at 37 6C in the phosphate buffer (pH= 6.8). Results Among 200 prepared extracts, Verbascum kermanensis , Rosa damascene, Rosmarinus officinalis , Levisticum officinale , Zataria multiflora Sanguisorba minor , Alhagi camelorum , Pistacia vera , Vaccinium arcto- staphylus, Zhumeria majdae, Alpinia officinarum , Salvadora persica, and Thymus serpyllum showed more than 50% inhibitory effect on the alpha glucosidase. Conclusion These active plants have -

Discovery of Antibacterial Dietary Spices That Target Antibiotic-Resistant Bacteria
Article Discovery of Antibacterial Dietary Spices That Target Antibiotic-Resistant Bacteria Dan Zhang 1, Ren-You Gan 1,*, Arakkaveettil Kabeer Farha 1, Gowoon Kim 1, Qiong-Qiong Yang 1, Xian-Ming Shi 1, Chun-Lei Shi 1, Qi-Xia Luo 2, Xue-Bin Xu 3, Hua-Bin Li 4 and Harold Corke 1,* 1 Department of Food Science & Technology, School of Agriculture and Biology, Shanghai Jiao Tong University, Shanghai 200240, China; [email protected] (D.Z.); [email protected] (A.K.F.); [email protected] (G.K.); [email protected] (Q.-Q.Y.); [email protected] (X.-M.S.); [email protected] (C.-L.S.). 2 State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, Zhejiang University, Hangzhou 310003, China; [email protected] 3 Department of Microbiology, Shanghai Municipal Center for Disease Control and Prevention, Shanghai 200336, China; [email protected] 4 Guangdong Provincial Key Laboratory of Food, Nutrition and Health, Guangdong Engineering Technology Research Center of Nutrition Translation, Department of Nutrition, School of Public Health, Sun Yat-Sen University, Guangzhou 510080, China. [email protected] * Correspondence: [email protected] (R.-Y.G.); [email protected] (H.C.); Tel.: +86-21-3420-8517 (R.-Y.G.); +86-21-3420-8515 (H.C.) Received: 11 April 2019; Accepted: 28 May 2019; Published: 29 May 2019 Abstract: Although spice extracts are well known to exhibit antibacterial properties, there is lack of a comprehensive evaluation of the antibacterial effect of spices against antibiotic-resistant bacteria. -

Screening of Crude Drugs Used in Japanese Kampo Formulas for Autophagy-Mediated Cell Survival of the Human Hepatocellular Carcinoma Cell Line
Medicines 2019, 6, 63; doi:10.3390/medicines6020063 S1 of S6 Supplementary Materials: Screening of Crude Drugs Used in Japanese Kampo Formulas for Autophagy-Mediated Cell Survival of the Human Hepatocellular Carcinoma Cell Line Shinya Okubo, Hisa Komori, Asuka Kuwahara, Tomoe Ohta, Yukihiro Shoyama and Takuhiro Uto Table S1. List of crude drugs. Drug Japanese Name English Name Scientific Name Medicinal Part No. 1 Akyo Donkey Glue Equus asinus glue 2 Ireisen Clematis Root Clematis chinensis, C. mandshurica, C. hexapetala root with rhizome 3 Inchinko Artemisia Capillaris Flower Artemisia capillaris capitulum 4 Uikyo Fennel Foeniculum vulgare fruit 5 Uzu a) Aconite Root Aconitum carmichaeli, A. japonicum tuberous root (mother root) 6 Uyaku Lindera Root Lindera strychnifolia root 7 Engosaku Corydalis Tuber Corydalis turtschaninovii tuber 8 Ogi Astragalus Root Astragalus membranaceus, A. mongholicus root 9 Ogon Scutellaria Root Scutellaria baicalensis root 10 Obaku Phellodendron Bark Phellodendron amurense, P. chinense bark 11 Oren Coptis Rhizome Coptis japonica, C. chinensis, C. deltoidea, C. teeta rhizome 12 Onji Polygala Root Polygala tenuifolia root or root bark 13 Gaiyo Artemisia Leaf Artemisia princeps, A. montana leaf and twig 14 Kashi Myrobalan Fruit Terminalia chebula fruit 15 Kashu Polygonum Root Polygonum multiflorum root 16 Gajutsu Zedoary Curcuma zedoaria rhizome 17 Kakko Pogostemon Herb Pogostemon cablin aerial part 18 Kakkon Pueraria Root Pueraria lobata root 19 Kasseki Aluminum Silicate Hydrate with Silicon Dioxide 20 Karokon Trichosanthes Root Trichosanthes kirilowii, T. kirilowii var. japonica, T. bracteata root Medicines 2019, 6, 63; doi:10.3390/medicines6020063 S2 of S6 21 Karonin Trichosanthes Seed Trichosanthes kirilowii, T. kirilowii var. japonica, T. -

A Review on the Ethnomedicinal Uses, Phytochemistry and Pharmacology of Plant Species Belonging to Kaempferia L
Pharmaceutical Sciences Asia Pharm Sci Asia 2021; 48(1), 1-24 DOI:10.29090/psa.2021.01.19.070 Review A review on the ethnomedicinal uses, phytochemistry and pharmacology of plant species belonging to Kaempferia L. genus (Zingiberaceae) Ngoc Khanh Pham1,2,3*, Hoang Tuan Nguyen2, Quoc Binh Nguyen4 ABSTRACT 1 Institute of Natural Products Chemistry Kaempferia L. is a genus commonly distributed in Asian (INPC), Vietnam Academy of Science and Technology, Cau Giay, Hanoi, Vietnam countries including China, India, Thailand, Myanmar, Malaysia, 2 Hanoi University of Pharmacy, Hoan Kiem, Indonesia, Laos, Cambodia and Vietnam, where these species are Hanoi, Vietnam popularly used as traditional medicines for different ailments 3 College of Pharmacy, Dongguk University, Goyang, Korea comprising infective diseases, wound infection, cough, pain and 4 Vietnam National Museum of Nature, digestion disorders on chemical composition of Kaempferia plants Vietnam Academy of Science and revealed the presence of natural compounds classified in Technology, Cau Giay, Hanoi, Vietnam monoterpenoids, diterpenoids, flavonoids, phenolic glycosides, cyclohexane oxide derivatives, diarylheptanoids and essential oil with *Corresponding author: various biological properties, which are valuable for discovery of new Ngoc Khanh Pham natural-derived therapeutic drugs and applications for the human [email protected] beings. This study is aimed to review the chemical, ethnobotanical and pharmacological properties of the plants belonging to Kaempferia genus growing in Asian countries and especially in South East Asia. 1. INTRODUCTION Kaempferia, is a medium - sized genus of about 60 plant species belonging to Zingiberaceae1 that is one of the major tropical plant families with many members commonly used as ornaments, 2 spices and as medicinal herbs . -

Edited Perennials List Spring 2019
2020 Nursery Season Perennials List Culinary Herbs Acorus calamus Sweet Flag Acorus gramineus 'Pusillus Minimus Aureus' Dwarf Golden Sweet Flag Acorus gramineus variegatus Grassy Sweet Flag Alpinia galanga Greater Galangal Alpinia officinarum Lesser Galangal Armoracia rusticana Horseradish Artemisia dracunculus French Tarragon Cryptotaenia japonica Mitsuba Cymbopogon flexuosus East Indian Lemongrass Eriocephalus africanus African Rosemary Hyssopus officinalis Hyssop Blue-Flowered Hyssopus officinalis Hyssop Pink-Flowered Hyssopus officinalis Hyssop White-Flowered Micromeria fruiticosa White Savory Pelargonium crispum Golen Lemon Crisp Geranium Pelargonium 'Attar of Rose' Rose Geranium Pelargonium fragrans Candy Dancer Pelargonium sp. Nutmeg Geranium Pelargonium sp. Nutmeg Variegated Polygonum odoratum Vietnamese Cilantro Sanguisorba minor Salad Burnet Satureja montana Winter Savory Satureja spinosa Pygmy Savory Satureja thymbra Savory of Crete Silene inflata Stridolo/Sculpit Smyrnium olusatrum Alexanders Stevia rebaudiana Stevia Zingiber mioga Japanese Mioga Ginger Zingiber mioga variegata Japanese Ginger 'Dancing Crane' Culinary Herbs & Edible Flowers Agastache foeniculum Blue Anise Hyssop Agastache foeniculum White Anise Hyssop Allium schoenoprasum Chives Allium tuberosum Garlic Chives Levisticum officinale Lovage Tulbaghia violacea White Flowered Society Garlic Tulbaghia violacea Society Garlic Tulbaghia violacea Variegated Society Garlic Selection subject to change, while supplies last. Questions? Call the nursery at (707) 874-9591. -

Bioactivity of Two Extracts from Alpinia Officinarum Rhizome Against Tribolium Castaneum (Herbst) Adults
African Journal of Biotechnology Vol. 11(16), pp. 3788-3792, 23 February, 2012 Available online at http://www.academicjournals.org/AJB DOI: 10.5897/AJB11.3847 ISSN 1684–5315 © 2012 Academic Journals Full Length Research Paper Bioactivity of two extracts from Alpinia officinarum rhizome against Tribolium castaneum (Herbst) adults Jianhua Lü 1*, Xiali Guo 2 and Lailin Zhang 1 1School of Food Science and Technology, Henan University of Technology, Zhengzhou 450052, China. 2School of Chemical Engineering and Energy, Zhengzhou University, Zhengzhou 450001, China. Accepted 1 February, 2012 The bioactivities of two solvent plant extracts prepared from Alpinia officinarum Hance rhizome, a traditional Chinese herbal plant were investigated against Tribolium castaneum (Herbst) adults in the laboratory. The rhizome extracts were prepared by n-hexane and methylene chloride extraction. Both extracts had significant repellent activity against T. castaneum adults with over 89% at class V at the dosage of 0.10 µl/cm 2 and above after 24 h exposure. The repellent activity significantly increased with increased exposure dosage. Both extracts also had strong fumigant activity against T. castaneum adults with over 80.0% mortality at the highest dosage tested of 80 µl/l air after 48 h of exposure. Mortality was dose dependent for both extracts. Regardless of extraction method A. officinarum rhizome extracts were demonstrated to have bioactivity against T. castaneum adults. These results suggest that the A. officinarum rhizome extracts have potential for integrated pest management programs of T. castaneum population. Key words: Alpinia officinarum rhizome extract, Tribolium castaneum (Herbst), fumigant activity, repellent activity, plant extract. INTRODUCTION The red flour beetle, Tribolium castaneum (Herbst) ancient times in China and abroad. -

Research Paper Principal Component and Heat Map Analysis of The
Academia Journal of Medicinal Plants 6(6): 101-113, June 2018 DOI: 10.15413/ajmp.2018.0122 ISSN 2315-7720 ©2018 Academia Publishing Research Paper Principal component and heat map analysis of the biological activities for some selected medicinal plants Accepted 8th June, 2018 ABSTRACT African and Asian countries have a gorgeous and prestigious heritage of herbal medicines. Ten well-known medicinal plants cultivated and grown under African environmental conditions (temperature, relative humidity, irradiance, photoperiod, wind and soil properties, etc) were extracted with methanol. Themethanolic extracts of these plants were screened for their antileishmanial, cytotoxic and free radical scavenging activities. The methanol extract of Colchicum autumnale and Alpinia officinarum showed high antileishmanial activity (IC5061.85±2.09 and 66.42±1.60 µg/ml, respectively). A. officinarum exhibited the most potent cytotoxicity (IC50 8.66±2.30 and 3.66±1.52 µg/ml against Hep G2 and A549, respectively). Rosa damascena, A. officinarum, and Humulus lupulus showed potent DPPH radical scavenging activity with IC50 values (2.0±0.23, 11.50±2.17, 14.0±2.0 µg/ml, respectively). Principal component analysis (PCA) was used as a Ahmed G. G. Darwish1,2*, Mamdouh N. statistical tool for experimental sets of obtained values for antileishmanial, Samy2,3, Sachiko Sugimoto2 Katsuyoshi antioxidant and anticancer activities performed statistics with person correlation Matsunami2 and Hideaki Otsuka2,4 matrix on plant samples. The results showed that there was a high correlation between anticancer activity against A549 and Hep G2 cells (0.743); moderate 1Department of Biochemistry, Faculty of Agriculture, Minia University, Minia correlation between anticancer activity against A549 cells and antileishmanial 61519, Egypt. -

Sweden Elixir
FLORA ORIGINAL SWEDEN ELIXIR Its preparation took months or even years and there were closely guarded recipes sold in apothecaries to those who could afford them . In the 18th century, Swedish Bitters was rediscovered by Swedish medics Dr . Claus Samst and Dr . Urban Hjärne and in modern times it has been popularised by Maria Treben, an Austrian herbalist . This formula is not simply a collection of bitter herbs . It is the culmination of centuries of wisdom of experience with how different herbs interact with our digestive system . Flora’s Original Sweden Elixir combines herbs that have bitter laxative effects, anti-inflammatory and anti-spasmodic effects, cooling anti-microbial actions and warming, blood circulating effects for an overall tonic, strengthening result . From the mouth through the entire alimentary canal to the colon, Original Sweden Elixir exerts a positive influence . FLORAdvantage: available as: • Made in Canada • Unique, time-honoured formula from Europe Size Code UPC NPN • Alcoholic extract ensures the formula contains the 100 ml 8200 0 61998 08200 3 80038619 maximum active ingredients from the herbs – more potent than a water extraction 250 ml 8202 0 61998 08202 7 80038619 • Synergistic formula from centuries of traditional use Digestion 35 g 8201 0 61998 08201 0 80038634 ensures a balanced overall effect • Does not require refrigeration • Stimulates digestive juices and appetite NPN CLAIM to ensure nutrient absorption • Contains herbs traditionally used in Herbal • Mild laxative effect Medicine to relieve symptoms such as dyspepsia • Reduces gas and bloating (indigestion) and constipation. • Certified Kosher, non-GMO and vegan Ingredients and Dosage/Directions: Liquid: Description: Per 5 ml (1 tsp) contains: The bitter taste was part of the diet of many traditional MEDICINAL INGREDIENTS: people: it is included in the meals of the Jewish Passover, in Aloe (Aloe ferox leaf) . -

Ingwergewächse - Eine Ziemlich Scharfe Familie
DI Doris Lengauer Ingwergewächse - eine ziemlich scharfe Familie Der Boom der würzigen Gesellen in der Küche hat auch vor den Toren der Versuchsstati- on für Spezialkulturen nicht haltgemacht und so gab es bereits 2018 erste Tastversuche mit Ingwer und Curcuma, um deren Anbau in der Steiermark zu testen. Heuer beschlos- sen wir noch weitere Vertreter genauer unter die Lupe zu nehmen und, im Gegensatz zu vergangenem Jahr, nicht in Töpfen, sondern in Erde im Folientunnel zu kultivieren. Alle Ingwergewächse werden vegetativ über Rhi- zome vermehrt und sie erheben Anspruch an hohe Luftfeuchtigkeit während der gesamten Kulturdau- er. Die Anzucht von Ingwer und Curcuma erfolgte Anfang März. Es wurde Konsumware aus dem Le- bensmitteleinzelhandel für die Anzucht verwendet. Dafür wurden die Rhizome in Teilstücke mit 2 Augen geteilt und in Substrat gelegt (siehe Foto rechts). Von den anderen Gattungen wurden Jungpflanzen erworben. Die Auspflanzung erfolgte im Mai im Fo- Die Ingwerrhizome wurden in 20 g Stücke mit 2 Augen zerteilt und in ein Substrat gelegt. lientunnel mit einer Pflanzendichte von 4 Pflanzen am Quadratmeter. Galgant gelangte im Mittelalter nach Europa. Hilde- gard von Bingen berichtete bereits im 12. Jahrhun- Galgant (Alpina officinarum ) Synonyme: dert über die Anwendung bei Magen- und Darmer- „kleiner“ Galgant, Echter Galgant, Siam Ingwer krankungen. Die Droge wurde bereits als Gewürz Hauptanbaugebiete: verwendet. Mittlerweile ist auch eine fungizide, Thailand, China, Japan, Indien tumor- und phagozytosehemmende Wirkung nach- gewiesen. Ähnlich wie Ingwer dient Galgant als Gewürz zahlreicher Fleisch-, Fisch- und Süßspeisen und ist beispiels- weise im berühmten indonesischen Gericht „Nasi Goreg“ verantwortlich für den typischen Geschmack. Ex- trakte aus Galgant werden außer- Galgant Pulver dem als Zusatz zu Likören (Magen- bitter), in Skandinavien und Rußland auch für Bier verwendet.