16S Rdna Based Identification from Highly Polluted Bangalore, India NA
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Lvo-45A Rc Details-123
ÀЦÑÍ´ ÌБÐÔÜ ƒ¸°º¦ÐÔÀÐÔ 2005¤Ð »Ðõ‘Фб ‘Ðà 4 (1)(Š) ªÁ.vÉ.¸À.D, ¸ÀܽÃAiÀÄ ªÀiË®åªÀ¢üðvÀ vÉjUÉ PÀbÉÃj-045(C¥ÀgÀ), ¨ÉAUÀ¼ÀÆgÀÄ. Details of RC Records in the office of ACCT, LVO-045(Addl.), Bangalore as on 30-09-2011. ‘ЮгзРÀЗÓþ ‘ЮгÐÀйÐÔî ‘Ð®Ð³ÐÀйÐÔî ‘Ð®Ð³ÐÀйÐÔî ‘Ð®Ð³Ð ‘ЮгР‘ЮгР‘ЮгР¹ÑÆÐ ÁÈЦÐÔÁÈЦÐÔ ÁÈЦÐÔ ‘ЮгР‘ЮгР‘ЮгР‘ЮгРƒ³Ðô ‘ЮгРºÀÐþÌв٠‘ЮгР‘ЮгР‘ЮгБЮгР‘ЮгР‘ЮгР‘ÐõÀÐÔ ‘ÐõÀÐÔ ‘ÐõÀÐÔ·Ñ“ÄÑ´–ÐÎÐ ‘ЮгР‘Фб ƒ¿°ÄÙÓ”ÑæÐÔ ƒ¿°ÄÙÓ”ÑæÐÔ‘ÙÜ ƒ¿°ÄÙÓ”ÑæРƒ¿°ÄÙÓ”Ñ Ã¦ÐÔ·Ð ¹ÑÆЖÙÖÏÊÐÃԻЯ˷Р»Ð¯Ë·Ð »Ð¯Ë·Ð »Ñõ¤Ð¿°Ë·Ð –ÙÖÏË·Ð ÁÄÙÓÀѧ ¹ÑÆЖÙÖÏË· ÊДÙôÊДÙô ÊДÙôÊДÙôÊДÙô ÊДÙô Š ¿ Ë ·ÐÅö ‘ÐÎÐÔÍË·Ð ‘ÐÎÐÔÍ˷Р˽ñ¸ Ô·ÐÅö »Ð®Ù·Ð ƒ¸°‘ѧ ÌÙÊФÐÔ „·ÙÓÇ˷Ѓ¸°‘ѧ ƒ¸°‘ѧ ƒ¸°‘ѧ ÀÐÈÐþÀÐÈÐþ ÀÐÈÐþ¸¹Ñ‘и¹Ñ‘Ð ¸¹Ñ‘Ð ÀÐÈÐþÀÐÈÐþÐ ¸¹Ñ‘Ð ÀÐÈÐþ Ð ¸¹Ñ‘Ð Ð ¸¹Ñ‘Ð ¯ …¸¹Ñ‘и¹Ñ‘Ð ¸¹Ñ‘ÐÌÙÊФÐÔÌÙÊФÐÔ ÌÙÊФÐÔ¸¹Ñ‘и¹Ñ‘Ð ¸¹Ñ‘Ð ƒ¸°‘ѧ ÌÙÊФÐÔÌÙÊФÐÔÌÙÊФÐÔ ÌÙÊФÐÔ ¤Ñô‘ý ¹Ð¤Ñô‘ý ¹Ð ¤Ñô‘ý ¹Ð½®ÐÄý ¹Ð.ÀÐÈÐþÀÐÈÐþ ÀÐÈÐþ 1 2 3(1) 3(2) 4 5 6 7 8 9 10 11a 11b 11c 12 13 14 15 GROUP C NO. -

Storename Address Ramamurthy Nagar Vodafone Store,Nithin
Storename Address Vodafone Store,Nithin Arcade, No 8, Ramamurthy Nagar, Banaswadi Main road, Bangalore Ramamurthy Nagar – 560043 Vodafone Store, Ground floor, No.35, 1st Main Road, Opp to Commercial Tax Office, Gandhinagar Gandhinagar, Bengaluru-560009 Vodafone Store, c/o, Maruthi clothing Co., Plot No.7-D, 1st phase,Doddanekundi industrial Whitefield area, Whitefield road, Bengaluru-560048 Vodafone Store, G-06 Lower ground floor, Innovater Building, concorase mall, Itpl International Tech park, Bengaluru-560066 Bommanahalli Vodafone store No.75/32/2, Begur main Road, Bommanahaali, Bangalore-560068 Vodafone Store, Ground Floor, CJR Complex, Outer Ring Road, Bellandur, Near Sarjapur Bellandur road junction, Opp to Café Coffee Day, Bengaluru-560 037. Vodafone Store, No 90, Sighmond Towers, Marthahalli Outer Ring Road, Bangalore – Marthahalli 566037 Hsr Layout Vodafone Store, No 806,27th Main, 80 Feet Road, HSR Layout Sector 1, Bengaluru-560102 Vodafone Store, Mehta Arcade, No.71, 15th Cross, 100 Feet Road, JP Nagar 6th Phase, Jp Nagar Bengaluru-560076 Vodafone Store, #113, Prestige Pinnacle, Koramangala Industrial Estate, Opp. Raheja Koramangala Arcade, Koramangala, Bengaluru-560095 Vodafone Store, No65/1A, Opp to Bata Showroom, Kaikondanhalli, Sarjapur Main Road, SARJAPUR ROAD Bangalore - 35 Vodafone Store, No Suraj Towers, Ground Floor, 27th cross, 3rd block, Jayanagar, Jayanagar bangalore-560011 Infosys-Blr Vodafone Store, Infosys technologies Ltd., Plot no. 40, Electronic city, Bengaluru-560100 Vodafone Store,No 21/A, 7th Main, 80 Feet Road, 1st Block, Koramangala, Bangalore Koramangala 80 feet road 560034 Vodafone Store,No 62-B, Majestic Terraces, ground floor, electronic city phase-1, Electronic City Bangalore - 560100 Vodafone Store, No 565,30th Main Road,DG Petrol Bunk Road,Banagiri Nagar,Next To Banshankari Mega Mart ,Bengaluru-560085 Vodafone Store, No. -

Ltd., Cambridge Road, Bangalore
List of IT Companies registered with Department and availed Power Tariff Concession Certificate(PTCC) SL No Compoany Name and address 2002-03 M/s. Synova Innovatinve Technologies (P) Ltd., 1 Cambridge Road, Bangalore - 8 M/s. Trivium India Software (P) Ltd, 138/6, 6th A Cross RMV Extension, Sadashivanagar, 2 Bangalore. M/s. A.K. Aerotak Software Centre (P) Ltd. 3 No.1, HAL II 'A' Stage, Bangalore - 8 M/s. Rational Software Corporation (India) Pvt. Ltd. No. 3K, Esteem Asrain, Koramangala Industrial Layout, 4 Bangalore - 34 2003-04 M/s. ICICI Infotech Ltd. "Brigade Champak", No. 6/2, Union Street, Off: Infantry 5 Road, Bangalore - 01 M/s. L.G. Soft India (P) Ltd. 5th Floor, Tower 'B', Golf View Campus, Hind Tunnel Road, 6 Murugeshpalya, Bangalore - 17 M/s. GXS India Technologies Centre (P) Ltd., No. 841/1, 100 Ft. Road, Binnamangala, Indiranagar, 7 Bangalore M/s. Socrates Software India (P) Ltd. Prestige Atlanta, No. 10, Industrial Layout, III Block, 8 Koramangala, Bangalore - 560 034 2004-05 M/s. Satyam Computer Service Ltd. No. 44 (P), 45 (P), 46 (P), Electronic City Phase - II, Bangalore - 79 Amended Date: 04-06-2015 Change of company name 9 to M/s. Tech Mahindra Limited M/s. Hewlett Packard India, Software Operations (P) Ltd. Regd. Off. No. 29, Cunningham Road, Bangalore - 52 Having its Software Development Centres at the following address Unit No. 1 - No. 29, Cunningham Road, Bangalore - 1 Unit No. 2 - Express Building, No. 14c, Queens Road, Civil Station Bangalore - 1 Unit No. 3 - No. 30, Cunningham Road, Bangalore - 1 Unit No. -

To.No.ADM-I / 05 /2020 Office of the Chief Metropolitan Magistrate, Bengaluru, Date 26.05.2020
To.No.ADM-I / 05 /2020 Office of the Chief Metropolitan Magistrate, Bengaluru, Date 26.05.2020 SPECIAL NOTIFICATION Sub: Allocation of business to the newly established XXXVI, XXXVII, XXXVIII, XXXIX, XL & XLI ACMM Courts, Bengaluru –reg. Ref:- 1. G.O. Nos. LAW 137 LCE 2014 dated 26.03.2015 & LAW 137 LCE 2014 dated 28.03.2016 creating the Courts of XXXVI, XXXVII, XXXVIII, XXXIX, XL & XLI ACMM Courts, Bengaluru. 2. Posting of officers to the newly established 06 ACMM Courts vide Notification No. GOB (I)/4(2)/2020 dated 30.04.2020 of the Hon'ble High Court of Karnataka, Bengaluru. 3. Letter No. ADM-I(A)/251/2020 dated 15.05.2020 and ADM-I(A)/256/2020 dated 21.05.2020 of the Registrar, City Civil Court, Bengaluru. * * * In Consultation with the Hon'ble Principal City Civil and Sessions Judge, Bengaluru and in exercise of powers conferred under Section 19 (3) of Criminal Procedure Code 1973, I, ROOPA R. KULKARNI , Chief Metropolitan Magistrate, Bengaluru, do hereby issue this Special Notification regarding allocation of business to the newly established XXXVI, XXXVII, XXXVIII, XXXIX, XL & XLI ACMM Courts as hereunder; “The territorial jurisdiction of the Police Stations along with listed cases as per Annexure-C are hereby withdrawn from respective Courts and assigned to the newly established XXXVII, XXXIX & XLI ACMM Courts, as per Annexure-A”. “The territorial jurisdiction of the Police Stations along with listed cases as per Annexure-C are hereby withdrawn from the respective Courts and assigned to the newly established XXXVI, XXXVIII & XL ACMM Courts, as per Annexure-B”. -

(Lakes) in Urban Areas- a Case Study on Bellandur Lake of Bangalore Metropolitan City
IOSR Journal of Mechanical and Civil Engineering (IOSR-JMCE) e-ISSN: 2278-1684,p-ISSN: 2320-334X, Volume 7, Issue 3 (Jul. - Aug. 2013), PP 06-14 www.iosrjournals.org Scenario of Water Bodies (Lakes) In Urban Areas- A case study on Bellandur Lake of Bangalore Metropolitan city Ramesh. N 1, Krishnaiah. S2 1(Department of Civil Engineering, Government Engineering College, K.R.Pet-571 426, Karnataka) 2(Department of Civil Engineering, JNTUA College of Engineering, Anantapur -515 002, Andra pradesh) Abstract: Environment is made up of natural factors like air, water and land. Each and every human activities supports directly/indirectly by natural factors. India is facing a problem of natural resource scarcity, especially of water in view of population growth and economic development. Due to growth of Population, advancement in agriculture, urbanization and industrialization has made surface water pollution a great problem and decreased the availability of drinking water. Many parts of the world face such a scarcity of water. Lakes are important feature of the Earth’s landscape which are not only the source of precious water, but provide valuable habitats to plants and animals, moderate hydrological cycles, influence microclimate, enhance the aesthetic beauty of the landscape and extend many recreational opportunities to humankind .For issues, perspectives on pollution, restoration and management of Bellandur Lake Falls under Bangalore Metropolitan city is very essential to know their status but so far, there was no systematic environmental study carried out. Hence now the following studies are essential namely Characteristics, Status, Effects (on surrounding Groundwater, Soil, Humans health, Vegetables, Animals etc.,), resolving the issues of degradation, preparation of conceptual design for restoration and management. -

M/S Mckinley Ventures Sy No 76/2 ,76/3 Panathur Village ,Varthurhobli ,Bangalure
FORM – I & I A (Only for construction projects listed under item 8 of the Schedule) CONCEPTUAL PLAN & RELATED ANNEXURES FOR THE PROPOSED RESIDENTIAL BUILDING APARTMENT BY M/s Mckinley Ventures Sy no 76/2 ,76/3 Panathur Village ,varthurHobli ,Bangalure Prepared by M/s Mckinley Ventures #796, 9th ‘A’ Main, 9th A Main Rd, Binnamangala, Stage 2, Indiranagar, Bengaluru, Karnataka 560038. Table of Contents Contents Page No. Form 1 3 Form 1A 13 1 Location Map 27 2 Surrounding Features 28 3 Drawing of site Plan 29 4 Drawing of Basement Plan 30 5 Drawing of Ground Floor Plan 31 6 Drawing of Typical Floor Plan 32 7 Drawing of Overall Terrace Plan 33 8 Drawing of Elevation and Section 34 9 Details of Septic Tank & Soak Pit 35 10 Feasibility report on water pollution control 36 system 11 Rain Fall & Run Off calculations 47 12 Storm water Pollution Prevention Plan 49 13 Landscape Details 53 14 Traffic Impact Studies & Management 54 Measures 15 Impact of Noise & Mitigation Measures 56 16 Energy Conservation aspects in Building 56 materials 17 Objectives and Strategies of Fire Fighting 60 Design 18 Environmental Management Plan 62 19 Risk Assessment, Disaster Management 67 including Offsite & Onsite Emergency Plans 20 Conceptual Plan 86 Form 1 APPENDIX I (See paragraph – 6) Sl. No. Item Details 1 Name of the project/s Proposed residential building apartment by Mr., Vinod Kumar Reddy A M/s Mckinley Ventures Managing Partner #796, 9th ‘A’ Main, 9th A Main Rd, Binnamangala, Stage 2, Indiranagar, Bengaluru, Karnataka 560038 2 S. No. in the Schedule 8 3 Proposed capacity/area/length/tonnage Proposed residential building of 260 flats to be handled/ command area/lease with total built-up area of 47600 Sqmtrs area/number of wells to be drilled consisting of: BF+SF+13UF in total site area of 13463.95 Sq. -
Sl No District CVC Name Category 1 BBMP a D Halli UPHC Government
ಕ ೋ풿蓍 ಲಕಾಕರಣ ಕ ೋᲂ飍ರಗಳು (COVID VACCINATION CENTRES) Sl No District CVC Name Category 1 BBMP A D Halli UPHC Government 2 BBMP A M HOSPITAL Private 3 BBMP A Narayanapura PHC P3 Government 4 BBMP A V Multispeciality Hospital Private 5 BBMP A.V.HOSPITAL COVAXIN Private 6 BBMP Aaxis Hospitals P3 Private 7 BBMP AAYUG MULTISPECIALTY HOSP P3 Private 8 BBMP ABBIGERE UPHC 1 Government 9 BBMP ABHAY HOSPITAL Private 10 BBMP ABHAY HOSPITAL C1 Private 11 BBMP ABHAYA HOSPITAL Private 12 BBMP Abhayahasta Hospital Block 1 Private 13 BBMP ACURA HOSPITAL - COVAXIN Private 14 BBMP Acura Speciality Hospital Private 15 BBMP ADARSH HOSPITAL C1 Private 16 BBMP Adugodi Dispensary Government 17 BBMP Adugodi UPHC Government 18 BBMP Agadi HOSPITAL AND RESEARCH Private 19 BBMP Agara UPHC Government 20 BBMP Agrahara Dasarahalli UPHC Government 21 BBMP AGRAHARA LAYOUT UPHC Government 22 BBMP AKSHA HOSPITAL - JAYANAGAR Private 23 BBMP AMC Block 1 Private 24 BBMP AMC Block 2 Private 25 BBMP AMRUTAHALLI UPHC Government 26 BBMP ANANYA HOSPITAL 1 Private 27 BBMP Anjanappa Garden UPHC Government 28 BBMP Anjanapura UPHC Government 29 BBMP Anjanapura UPHC 1 Government 30 BBMP Apollo Clinic Doddakannelli P3 Private 31 BBMP APOLLO CLINIC HSR Private 32 BBMP Apollo Clinic Indiranagar Private 33 BBMP APOLLO CLINIC KORMANAGALA Private 34 BBMP Apollo Clinic Sarjapur P3 Private 35 BBMP APOLLO CM FERTILITY -JP NAGAR Private 36 BBMP APOLLO CRADLE - Koramangala Private 37 BBMP APOLLO CRADLE HOSPITAL Private 38 BBMP APOLLO CRADLE KLM - COVAXIN Private 39 BBMP Apollo Cradle P3 Private 40 BBMP APOLLO HOSPITAL 1 Private 41 BBMP APOLLO HOSPITAL BG Private 42 BBMP APOLLO HOSPITAL SHESHADRIPURAM Private 43 BBMP APOLLO HOSPITAL-C1 Private 44 BBMP Apollo Hospital-JAYANAGAR Private 45 BBMP APOLLO MEDICAL CENTER P3 Private 46 BBMP APOORVA Block 1 Private 47 BBMP APOORVA Hospital C1 Private 48 BBMP APTS UPHC Government 49 BBMP Arakere Uphc Government 50 BBMP Arka Hospital Private 51 BBMP ARTYEM HOSPITAL Private 52 BBMP ARTYEM HOSPITAL C1 Private 53 BBMP ARYAN MULTISPECIALITY HOS. -

Bellandur Development Forum Email: [email protected] Twitter: Facebook:
Bellandur Development Forum eMail: [email protected] Twitter: https://twitter.com/kdevforum facebook:https://www.facebook.com/groups/bellandurforum Date: 19-Oct-2019 To, Mr. Venkata Chalapathy, Joint Commissioner, Mahadevapura Zone Bangalore Subject: Commercial Building Violations without Parking & In Residential Areas Dear Sir, We are a forum in Bellandur (Ward 150, Bengaluru) and a citizen advocacy group representative of over 90 RWAs across Bellandur. Over the past few years, there is unrestricted growth happening especially of Commercial and Paying Guest Complex in Mahadevapura ITCorridor due to unplanned growth and non administration. We feel there is a clear collaboration between the BBMP, Town planning BDA officials and real estate, small time builders in this violation as none of sahaaya complaints, ward committee escalations or personal escalation through MLA offices are acted upon. We request : 1. A thorough investigation into each case the alleged nexus between Town planning, BDA and BBMP officials(AEE, AEs) with the Real Estate on the ground. We will soon represent a petition to UpaLokayukta too but wanted to highlight this issue. If you see tabular list below some are pending for action since 2017. 2. The table below, refer #1 through #13 have got BBMP notice issuance through former Joint Commissioner, Mahadevapura, BBMP but later no action by any BBMP officials in spite of reminding in-front of the same JC, AEE, CE. In one case(Refer#13 below) even Police FIR lodged with Bellandur Police Station from BBMP has been compromised. The concerned openly challenge citing they have took care of BBMP or concerned officials, so how can anyone evict them. -

Evaluation of Water Quality in Bellandur Lake
International Journal of Engineering Technology Science and Research IJETSR www.ijetsr.com ISSN 2394 – 3386 Volume 5, Issue 1 January 2018 Evaluation of Water Quality in Bellandur Lake Parvathi K S Dayananda Sagar College of Engineering, Bengaluru Shivani P Kumar Dayananda Sagar College of Engineering, Bengaluru Vikash Kumar Gupta Dayananda Sagar College of Engineering, Bengaluru ABSTRACT: Bangalore, a garden city has plenty of fresh water lakes and greenery around it. The lakes of the city are facing extinction due to pollution. One such highly polluted lake is 'Bellandur Lake’. Located at 18 km towards the south of the city with a water spread area of 2.5 sq. km. The lake catchment has been subjected to extreme environmental stress mainly due to lush unplanned developmental activities in the catchment in recent years. The development plans of the region ignore the integrated planning approaches considering all the components of the ecosystem. This lake is scattered throughout the region, therefore different samples were taken and detailed analysis were conducted Keywords: status of water quality, the physical and chemical characteristics of water INTRODUCTION: Water is one of the most significant natural resource available to mankind. Knowing the importance of water for sustenance of life, the need for conservation of water bodies especially the fresh water bodies is being realized worldwide. Lakes in Bangalore, Karnataka are numerous, and there are no rivers close by. Most lakes in the Bangalore region were constructed in the sixteenth century by damming the natural valley systems by constructing bunds. In recent years, the Management of Lakes traditionally done by the government agencies witnessed experimentation by the Lake Development Authority with a limited public–private sector participation in respect of three lakes, which has proved controversial and resulted in almost a reversal of the policy. -

Bangalore Urban Ward Details
Ward Name District Taluka Hobli Ward Number Ward Name in English Area Comes Under Concern Ward in Kannada Govindapura, Kulappa Layout, Vasudevapura, Kendriya Vihar, Manchenahalli, Yelahanka (P), Yelhanka Airport Area, Maheshwari Nagar, Sai spring field colony, Gandhi Nagar, Lake view residency, Nehru Nagar, Venkatala, Surabhi Layout, Venkatappa Layout, Shobha Ultima villas, Bangalore Bangalore North Addl Yelahanka-1 Ward-1 Kempegowda ಂಡ Venkatala Layout, Mantri township, Vikas layout, Yelahanka Kere, Shankaranna Layout, Sathyappa Anjanappa Kempamma Layout, Shivanahalli (P), Maruthi Nagar, Sapthagiri Layout (P), Jayanna Layout, Basaveshwara Nagar (P), Bhadranna Layout Harohalli, Harohalli kere, Kanchenahalli, ISRO Layout, Naganahalli, Naganahalli new layout, KEB Layout Phase I, Balaji Layout, Vinayaka Layout, Ramanashree Califonia East Garden layout, Deo Marvel Layout, Mahalaxmi Bangalore Bangalore North Addl Yelahanka-1 Ward-2 Chowdeshwari ಶ Layout, Nisarga Layout, CRPF Quarters, Puttanahalli, Puttanahalli kere, Monte Carlo apartment, DG staff quarters, Central excise quarters, Wheel and Axle plant, FM Goetze plant, Chowdeswari Layout, Kamakshiamma Layout, East Colony, Yelahanka (P), KHB Colony Ananthapura, Chikka Bettahalli, Dodda Bettahalli, Bharat Nagar (MS Palya), Chandrappa Layout, Hill side meadows layout, Sai Nagar Phase I and II, Basavalingappa Layout, Netravathi Layout (P), Sai orchards, Best country 3, G Ramaiah Layout, Jyothi Nagar, GPF Layout, Muneshwara Layout 1st and 2nd Bangalore Bangalore North Addl Yelahanka-3 Ward-3 -
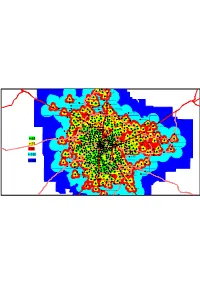
Coverage Plot of 3G Service in Bangalore City
KHB Colony - Yelahanka Attur L/O Yelahanka TE Yelehanka-1 Tumkur Road Vidyaranyaoura TE Chickbanavara MS Palya - Vidyaranyapura Allalasandra Judges L/O Bagalaguntai Abbigere thindlu Amuruthhalli Chikkasandra - Hesaraghatta Main Road Indian Express LO Madanayakanahalli Doddabommasandra KodigehalliGate,BellaryRoad Tumk_Rd_Jindal2 (BIEC) Hessaraghatta Main Road Jalhalli(Gangamma Circle) Bhuvaneswari Nagar Dasarahalli_TUMK_Rd (MS Ramaiah Nagar) Jindal Nagar Peenya Dasrahalli Bhadrappa L/O Hennur Main Road 2 Ayyappan Temple, Jalahalli Gokul Sanjaynagar TE Cholanayakanahalli Nagawara Junction HMT Colony, Neelagaddana halli Hebbal (Airtel Tower Building) Whitefield Road AG'S L/O RMV Hennur Cross Anand Nagar-II Nagavara Kelkere Village Peenya - SRS Hennur LIC Colony Anand Nagar Mallappa L/O Peenya I Stage Subedar Palya HBR L/O RSU Ganga Nagar - II Anand nagar, KR Puram M.S.Ramaiah Road Akshay Nagar PIE Goraguntapalya Ambedakar Medical College 2 Kalyan Nagar Thambu Chettipalya - KR Puram Nandini Layout Kavalbyrasandra-II Hoysala nagar BHEL, opp to IISc Ganapathi nagar Nandini L/O TE DJ Halli Banaswadi Malleshwaram 8th Main Lingarajapuram-II K.R Puram Market Hegganahalli-II Laggere-2 kurubarahalli-II Chowdaiah Memorial Hall Kasturinagar II Rajajinagar R Block CACT Tel exge Hegganahalli Kurubarahalli GuttaHalli Jayamahal Banasawadi Rly Stn Rd Ayyappanagar Basaveshwaranagar, Modi Hospital Kadugodi Cox Town Palace Road Benson Town Benniganahalli K R Puram RSU Basaveshwaranagar RSU Hoodi Frazer Town - III Basaveshwaranagar - III Block Seshadripuram -

COVID TEST ALGORITHM RT-PCR Test Rapid Antigen Test Antibody Tests (SARS- Cov2-Igg) Also Called Molecular Diagnostic Test Rapid Diagnostic Test Serology Tests
COVID TEST ALGORITHM RT-PCR Test Rapid Antigen Test Antibody Tests (SARS- CoV2-IgG) Also called Molecular diagnostic test Rapid diagnostic test Serology tests Confirmatory, when test For determining the % of results are positive. population or specific population group who Confirmatory Test (GOLD If test results are negative, have been infected in the Importance STANDARD) then a confirmatory RT- past and have recovered PCR Test needs to be without any symptoms or performed complications What it shows Current Infection Current Infection Past infection If you ever had COVID-19 If you ever had COVID-19 If you have an active What it doesn’t show in the past in the past COVID-19 infection Symptomatic persons Who can take it? Symptomatic persons Asymptomatic persons Should get tested once Post an infection During ongoing between day 5 and day with the virus, it symptoms 10 of coming into can take 2-3 weeks contacts with a to develop enough COVID19 positive antibodies to be OR as per doctor When to take it? patient. detected through advice an antibody test Or as per doctor advice Or as per doctor (or as per travel (or as per travel advice requirements) requirements) Follow-up tests (if no fever for 3 days after Generally Not required end of home isolation (Pls take your doctor’s Rarely Sometimes or hospital advice) discharge) Time to Get Test 24 to 48 Hours Upto 4 Hours Within 24 Hours Result Rs 500/- (in all branches of RxDx, except RxDx Rs 800 + *Rs 200 (*logistic Cost @ Clinic Rs 700/- SAMANVAY, charges) = Rs 1000 Malleswaram) COVID