Two Pectate Lyases from Caldicellulosiruptor Bescii with the Same CALG Domain Had
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

ATP-Citrate Lyase Has an Essential Role in Cytosolic Acetyl-Coa Production in Arabidopsis Beth Leann Fatland Iowa State University
Iowa State University Capstones, Theses and Retrospective Theses and Dissertations Dissertations 2002 ATP-citrate lyase has an essential role in cytosolic acetyl-CoA production in Arabidopsis Beth LeAnn Fatland Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/rtd Part of the Molecular Biology Commons, and the Plant Sciences Commons Recommended Citation Fatland, Beth LeAnn, "ATP-citrate lyase has an essential role in cytosolic acetyl-CoA production in Arabidopsis " (2002). Retrospective Theses and Dissertations. 1218. https://lib.dr.iastate.edu/rtd/1218 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Retrospective Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. ATP-citrate lyase has an essential role in cytosolic acetyl-CoA production in Arabidopsis by Beth LeAnn Fatland A dissertation submitted to the graduate faculty in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Major: Plant Physiology Program of Study Committee: Eve Syrkin Wurtele (Major Professor) James Colbert Harry Homer Basil Nikolau Martin Spalding Iowa State University Ames, Iowa 2002 UMI Number: 3158393 INFORMATION TO USERS The quality of this reproduction is dependent upon the quality of the copy submitted. Broken or indistinct print, colored or poor quality illustrations and photographs, print bleed-through, substandard margins, and improper alignment can adversely affect reproduction. In the unlikely event that the author did not send a complete manuscript and there are missing pages, these will be noted. -

The Genome of an Industrial Workhorse
NEWS AND VIEWS The genome of an industrial workhorse Dan Cullen Sequencing of the filamentous fungus Aspergillus niger offers new opportunities for the production of specialty chemicals and enzymes. Few microbes compare with the filamentous fungus Aspergillus niger in its ability to pro Environment CAT (2) H O H O + O duce prodigious amounts of useful chemicals 2 2 2 2 and enzymes. This fungus is the principal GOX (3) GLN (1) source of citric acid for food, beverages and D-glucono Glucose Gluconate Oxalate pharmaceuticals1 and of several important 1,5-lactone Citrate http://www.nature.com/naturebiotechnology http://www.nature.com/naturebiotechnology commercial enzymes, including glucoamy lase, which is widely used for the conversion of starch to food syrups and to fermentative Oxalate + acetate PEP feedstocks for ethanol production. Although OAH most of these fermentation processes are well cMDH (3) established, the underlying genetics are still cPYC (1) cACO (2) cIDH (1) poorly understood. In this issue, Pel et al.2 Pyruvate OAA MAL Citrate Isocitrate 2-ketoglutarate report the genome sequence of A. niger strain Cytosol CBS 513.88. The availability of this sequence OAT (1) CMC (2) mPYC (1) should provide invaluable aid toward improv Pyruvate OAA MAL Citrate Isocitrate 2-ketoglutarate ing the production of chemicals and enzymes PDH Nature Publishing Group Group Nature Publishing 7 in this organism. mMDH (1) mACO (2) mIDH (3) Pel et al. sequenced tiled bacterial artificial CS (3) 200 Acetyl-CoA TCA cycle © chromosomes representing the entire A. niger genome to produce a high-quality assembly of Mitochondrion 19 supercontigs with a combined length of 33.9 Mb. -

Structural Studies of Three Enzymes: Telomerase, the Methyltransferase Cobj and Pectate Lyase
Structural studies of three enzymes: Telomerase, the methyltransferase CobJ and Pectate lyase Teng Teng To Thesis submitted to the University of London for the Degree of Doctor of Philosophy 1 Abstract This thesis investigates the structure and function of three enzymes of biotechnological and biomedical interest: telomerase from Caenorhabtidis elegans , pectate lyase from Bacillus subtilis and the methyltransferase CobJ from Rhodobacter capsulatus . Telomerase is a ribonucleoprotein found in all eukaryotes and its function is to maintain telomere length, sustain chromosome integrity and circumvent the end-replication problem. The protein requires two subunits to function, telomerase reverse transcriptase (TERT), the catalytic component, and an intrinsic RNA template (TR). The TR makes telomerase a unique reverse transcriptase using the template in the synthesis of short iterative sequences which cap the ends of telomeres. This work reports the successful cloning of a small and therefore potentially crystallisable TERT from C. elegans and expression trials of this catalytic component. Cobalamin (vitamin B 12 ) is an intricate small molecule belonging to a group of compounds called cyclic tetrapyrroles. Its biosynthesis is achieved through a complex pathway encompassing over thirty different enzyme-mediated reactions. Within this pathway there are seven methyltransferases which add eight S-adenosyl-methionine (SAM) derived methyl groups to the macrocycle. CobJ catalyses the methylation of C17 and ring contraction at C20, this reaction which exudes C20 from the tetrapyrrole ring is unprecedented in nature. In this thesis I report the crystallisation of native CobJ and refinement and validation of a high resolution structure along side co-crystallisation and soaking experiments aimed at capturing an enzyme-tetrapyrrole complex. -

Pectate Lyase A, an Enzymatic Subunit of the Clostridium Cellulovorans Cellulosome
Pectate lyase A, an enzymatic subunit of the Clostridium cellulovorans cellulosome Yutaka Tamaru* and Roy H. Doi† Section of Molecular and Cellular Biology, University of California, Davis, CA 95616 Communicated by Arnold L. Demain, Massachusetts Institute of Technology, Cambridge, MA, January 29, 2001 (received for review August 2, 2000) Clostridium cellulovorans uses not only cellulose but also xylan, consisting of highly conserved 22-aa repeats. Therefore the C. mannan, pectin, and several other carbon sources for its growth cellulovorans cellulosomal enzymes identified to date are capable and produces an extracellular multienzyme complex called the of degrading cellulose, xylan, lichenan, and mannan. However, cellulosome, which is involved in plant cell wall degradation. Here because we have recently succeeded in converting Arabidopsis we report a gene for a cellulosomal subunit, pectate lyase A (PelA), and tobacco cells to protoplasts with the C. cellulovorans cellu- lying downstream of the engY gene, which codes for cellulosomal losomes (Y.T., S. Ui, H. Chan, R.H.D., and B. Liu, unpublished enzyme EngY. pelA is composed of an ORF of 2,742 bp and encodes data), and it has been reported that pectin could serve as a a protein of 914 aa with a molecular weight of 94,458. The amino carbon source for growth (5), we believed that the C. cellulo- acid sequence derived from pelA revealed a multidomain structure, vorans cellulosome must also have pectinase activity. The present i.e., an N-terminal domain partially homologous to the C terminus paper provides data that indicate that a cellulosomal gene pelA of PelB of Erwinia chrysanthemi belonging to family 1 of pectate and the enzyme encoded by this gene can degrade pectin and lyases, a putative cellulose-binding domain, a catalytic domain that pectate lyase A (PelA) contains a DS at its C terminus. -

Microbial Enzymes: Industrial Progress in 21St Century
3 Biotech (2016) 6:174 DOI 10.1007/s13205-016-0485-8 REVIEW ARTICLE Microbial enzymes: industrial progress in 21st century 1 1 2 3 Rajendra Singh • Manoj Kumar • Anshumali Mittal • Praveen Kumar Mehta Received: 8 April 2016 / Accepted: 1 August 2016 Ó The Author(s) 2016. This article is published with open access at Springerlink.com Abstract Biocatalytic potential of microorganisms have Introduction been employed for centuries to produce bread, wine, vinegar and other common products without understanding Microbes have been utilized since ancient human civi- the biochemical basis of their ingredients. Microbial lization with first reported commercial application of yeast enzymes have gained interest for their widespread uses in to produce alcoholic beverages from barley by the Baby- industries and medicine owing to their stability, catalytic lonians and Sumerians as early as 6000 BC. The microbial activity, and ease of production and optimization than plant enzymes have gained recognition globally for their wide- and animal enzymes. The use of enzymes in various spread uses in various sectors of industries, e.g., food, industries (e.g., food, agriculture, chemicals, and pharma- agriculture, chemicals, medicine, and energy. Enzyme ceuticals) is increasing rapidly due to reduced processing mediated processes are rapidly gaining interest because of time, low energy input, cost effectiveness, nontoxic and reduced process time, intake of low energy input, cost eco-friendly characteristics. Microbial enzymes are capable effective, nontoxic and eco-friendly characteristics (Li of degrading toxic chemical compounds of industrial and et al. 2012; Choi et al. 2015). Moreover, with the advent of domestic wastes (phenolic compounds, nitriles, amines recombinant DNA technology and protein engineering a etc.) either via degradation or conversion. -

Electronic Supplementary Information S10
Electronic Supplementary Material (ESI) for Metallomics. This journal is © The Royal Society of Chemistry 2019 Electronic Supplementary Information S10. Up and downregulated genes of ACR3 and TIP-ACR3 compared to control roots all exposed to 0.1 mM As III . HYBRIDIZATION 4: LIST OF UP-REGULATED GENES Fold Change Probe Set ID ([0.1 ACR3] vs [0.1-HR]) Blast2GO description Genbank Accessions C228_s_at 48.018467 N.tabacum cysteine-rich extensin-like protein-4 mRNA EB683071 C1359_at 32.31786 Proteinase inhibitor I3, Kunitz legume, Kunitz inhibitor ST1-like DW004832 EB430244_x_at 22.426174 unknow EB430244 C10896_at 21.463442 dir1 (defective in induced resistance 1) lipid binding JF275847.1 BP528597_at 21.445984 Mitochondrial DNA BP528597 C10933_x_at 20.347004 Solanum nigrum clone 82 organ-specific protein S2 (OS) EB443218 TT31_B05_s_at 17.260008 Proteinase inhibitor I3, Kunitz legume, Kunitz inhibitor ST1-like C3546_s_at 17.171844 fasciclin-like arabinogalactan protein 2 EB451563 C8455_at 16.855278 Defective in induced resistance 2 protein (DIR2) BP530866 C11687_at 16.618454 galactinol synthase EB432401 BP136836_s_at 15.364014 Nicotiana tabacum mitochondrial DNA BP136836 BP525701_at 15.137816 Nicotiana tabacum mitochondrial DNA BP525701 EB682942_at 14.943408 Hop-interacting protein THI101 EB682942 BP133164_at 14.644844 Nicotiana tabacum mitochondrial DNA BP133164 AY055111_at 14.570147 Nicotiana tabacum pathogenesis-related protein PR10a AY055111 TT08_C02_at 14.375496 BP526999_at 14.374481 Nicotiana tabacum mitochondrial DNA BP526999 CV017694_s_at -

Supplemental Table S1: Comparison of the Deleted Genes in the Genome-Reduced Strains
Supplemental Table S1: Comparison of the deleted genes in the genome-reduced strains Legend 1 Locus tag according to the reference genome sequence of B. subtilis 168 (NC_000964) Genes highlighted in blue have been deleted from the respective strains Genes highlighted in green have been inserted into the indicated strain, they are present in all following strains Regions highlighted in red could not be deleted as a unit Regions highlighted in orange were not deleted in the genome-reduced strains since their deletion resulted in severe growth defects Gene BSU_number 1 Function ∆6 IIG-Bs27-47-24 PG10 PS38 dnaA BSU00010 replication initiation protein dnaN BSU00020 DNA polymerase III (beta subunit), beta clamp yaaA BSU00030 unknown recF BSU00040 repair, recombination remB BSU00050 involved in the activation of biofilm matrix biosynthetic operons gyrB BSU00060 DNA-Gyrase (subunit B) gyrA BSU00070 DNA-Gyrase (subunit A) rrnO-16S- trnO-Ala- trnO-Ile- rrnO-23S- rrnO-5S yaaC BSU00080 unknown guaB BSU00090 IMP dehydrogenase dacA BSU00100 penicillin-binding protein 5*, D-alanyl-D-alanine carboxypeptidase pdxS BSU00110 pyridoxal-5'-phosphate synthase (synthase domain) pdxT BSU00120 pyridoxal-5'-phosphate synthase (glutaminase domain) serS BSU00130 seryl-tRNA-synthetase trnSL-Ser1 dck BSU00140 deoxyadenosin/deoxycytidine kinase dgk BSU00150 deoxyguanosine kinase yaaH BSU00160 general stress protein, survival of ethanol stress, SafA-dependent spore coat yaaI BSU00170 general stress protein, similar to isochorismatase yaaJ BSU00180 tRNA specific adenosine -

Evolving Strategies for Enzyme Engineering Jesse D Bloom1, Michelle M Meyer2, Peter Meinhold2, Christopher R Otey2, Derek Macmillan3 and Frances H Arnold1,2
Evolving strategies for enzyme engineering Jesse D Bloom1, Michelle M Meyer2, Peter Meinhold2, Christopher R Otey2, Derek MacMillan3 and Frances H Arnold1,2 Directed evolution is a common technique to engineer enzymes proteins with the desired enzymatic function. Such for a diverse set of applications. Structural information and an libraries can be designed by drawing on our knowledge understanding of how proteins respond to mutation and of how proteins respond to mutation [1–3] and of recombination are being used to develop improved directed sequence-structure-function relationships. These libraries evolution strategies by increasing the probability that mutant themselves in turn generate new information about pro- sequences have the desired properties. Strategies that target teins and protein evolution [4,5]. Here, we review recent mutagenesis to particular regions of a protein or use successes in the directed evolution of enzymes, with a recombination to introduce large sequence changes can special focus on how knowledge is incorporated into complement full-gene random mutagenesis and pave the way directed evolution strategies. Other recent reviews to achieving ever more ambitious enzyme engineering goals. describe in detail how these engineered enzymes have Addresses been utilized in chemical synthesis [6] and as components 1 Division of Chemistry and Chemical Engineering, Mail Code 210-41, of engineered pathways [7,8]. California Institute of Technology, 1200 East California Boulevard, Pasadena, CA 91125, USA Directed evolution strategies 2 Biochemistry and Molecular Biophysics, Mail Code 210-41, California Institute of Technology, 1200 East California Boulevard, Directed evolution works when the researcher can find at Pasadena, CA 91125, USA least one enzyme with improved properties in the 3 School of Chemistry, University of Edinburgh, King’s Building, sequence library. -

A Hierarchical Classification of Polysaccharide Lyases for Glycogenomics Vincent Lombard, Thomas Bernard, Corinne Rancurel, Harry Brumer, Pedro M
A hierarchical classification of polysaccharide lyases for glycogenomics Vincent Lombard, Thomas Bernard, Corinne Rancurel, Harry Brumer, Pedro M. Coutinho, Bernard Henrissat To cite this version: Vincent Lombard, Thomas Bernard, Corinne Rancurel, Harry Brumer, Pedro M. Coutinho, et al.. A hierarchical classification of polysaccharide lyases for glycogenomics. Biochemical Journal, Portland Press, 2010, 432 (3), pp.437-444. 10.1042/BJ20101185. hal-00539724 HAL Id: hal-00539724 https://hal.archives-ouvertes.fr/hal-00539724 Submitted on 25 Nov 2010 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Biochemical Journal Immediate Publication. Published on 07 Oct 2010 as manuscript BJ20101185 A hierarchical classification of polysaccharide lyases for glycogenomics V. Lombard*, T. Bernard*†, C. Rancurel*, H Brumer‡, P.M. Coutinho* & B. Henrissat*1 *Architecture et Fonction des Macromolécules Biologiques, UMR6098, CNRS, Université de la Méditerranée, Université de Provence, Case 932, 163 Avenue de Luminy, 13288 Marseille cedex 9, France ‡School of Biotechnology, Royal Institute of Technology (KTH), AlbaNova University Centre, 106 91 Stockholm, Sweden † Present address: Biométrie et Biologie Évolutive, UMR CNRS 5558, UCB Lyon 1, Bât. Grégor Mendel, 43 bd du 11 novembre 1918, 69622 Villeurbanne cedex, France 1To whom correspondence should be addressed: [email protected]‐mrs.fr Abstract: Carbohydrate‐active enzymes face huge substrate diversity in a highly selective manner with only a limited number of available folds. -
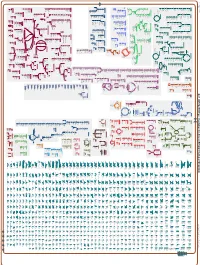
Generated by SRI International Pathway Tools Version 25.0, Authors S
Authors: Pallavi Subhraveti Ron Caspi Peter Midford Peter D Karp An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Ingrid Keseler Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_003722335Cyc: Caulobacter flavus RHGG3 Cellular Overview Connections between pathways are omitted for legibility. -

The Complete Genome Sequence of the Gram-Positive Bacterium Bacillus Subtilis
articles The complete genome sequence of the Gram-positive bacterium Bacillus subtilis F. Kunst1, N. Ogasawara2, I. Moszer3, A. M. Albertini4, G. Alloni4, V. Azevedo5, M. G. Bertero3,4, P. Bessie` res5, A. Bolotin5, S. Borchert6, R. Borriss7, L. Boursier3, A. Brans8, M. Braun9, S. C. Brignell10,S.Bron11, S. Brouillet3,12, C. V. Bruschi13, B. Caldwell14, V. Capuano5, N. M. Carter10, S.-K. Choi15, J.-J. Codani16, I. F. Connerton17, N. J. Cummings17, R. A. Daniel18, F. Denizot19, K. M. Devine20,A.Du¨sterho¨ ft9, S. D. Ehrlich5, P.T. Emmerson21, K. D. Entian6, J. Errington18, C. Fabret19, E. Ferrari14, D. Foulger18, C. Fritz9, M. Fujita22, Y.Fujita23,S.Fuma24, A. Galizzi4, N. Galleron5, S.-Y.Ghim15, P.Glaser3, A. Goffeau25, E. J. Golightly26, G. Grandi27, G. Guiseppi19,B.J.Guy10, K. Haga28, J. Haiech19, C. R. Harwood10,A.He´naut29, H. Hilbert9, S. Holsappel11, S. Hosono30, M.-F. Hullo3, M. Itaya31, L. Jones32, B. Joris8, D. Karamata33, Y.Kasahara2, M. Klaerr-Blanchard3, C. Klein6, Y.Kobayashi30, P.Koetter6, G. Koningstein34, S. Krogh20, M. Kumano24, K. Kurita24, A. Lapidus5, S. Lardinois8, J. Lauber9, V. Lazarevic33, S.-M. Lee35, A. Levine36, H. Liu28, S. Masuda30, C. Maue¨ l33,C.Me´digue3,12, N. Medina36, R. P. Mellado37, M. Mizuno30, D. Moestl9, S. Nakai2, M. Noback11, D. Noone20, M. O’Reilly20, K. Ogawa24, A. Ogiwara38, B. Oudega34, S.-H. Park15, V. Parro37,T.M.Pohl39, D. Portetelle40, S. Porwollik7, A. M. Prescott18, E. Presecan3, P. Pujic5, B. Purnelle25, G. Rapoport1, M. Rey26, S. Reynolds33, M. Rieger41, C. Rivolta33, E. Rocha3,12,B.Roche36, M. -

Protein Engineering of the Aldoxime Dehydratase from Bacillus Sp. Oxb
www.nature.com/scientificreports OPEN Protein engineering of the aldoxime dehydratase from Bacillus sp. OxB‑1 based on a rational sequence alignment approach Keiko Oike1, Jens Sproß1, Daisuke Matsui2, Yasuhisa Asano2* & Harald Gröger1* Recently, the program INTMSAlign_HiSol for identifying aggregation hotspots in proteins only requiring secondary structure data was introduced. We explored the utility of this program further and applied it for engineering of the aldoxime dehydratase from Bacillus sp. OxB‑1. Towards this end, the efect of inverting the hydropathy at selected positions of the amino acid sequence on the enzymatic activity was studied leading to 60% of our constructed variants, which showed improved activity. In part, this activity increase can be rationalised by an improved heme incorporation of the variants. For example, a single mutation gave a 1.8 fold increased enzymatic activity and 30% improved absolute heme incorporation. Aldoxime dehydratases (Oxds; E.C. 4.99) are special heme containing lyases involved in the “aldoxime–nitrile- pathway” of microbes (Fig. 1). Tese enzymes catalyse the dehydration of aldoximes to nitriles, which are metabo- lised to carboxylic acids by nitrile hydratases and amidases or by nitrilases1–4. Te frst discovered enzyme of this class is the Oxd from Bacillus sp. OxB-1 (OxdB), which was isolated from soil1. Tis enzyme naturally catalyses the dehydration of E- and Z-phenylacetaldoxime (E- and Z-PAOx) to phenylacetonitrile (PAN). It was purifed as wild-type enzyme and expressed heterologously in Escherichia coli (E. coli)5,6. However, only a handful of these Oxds have been identifed, isolated and characterised yet7–13. Oxds can be applied for the synthesis of nitriles 13–20 and dinitriles 21, compound classes with diverse applica- tions in the chemical and pharmaceutical industry 22.