How the Cell Deals with DNA Nicks
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Introduction of Human Telomerase Reverse Transcriptase to Normal Human Fibroblasts Enhances DNA Repair Capacity
Vol. 10, 2551–2560, April 1, 2004 Clinical Cancer Research 2551 Introduction of Human Telomerase Reverse Transcriptase to Normal Human Fibroblasts Enhances DNA Repair Capacity Ki-Hyuk Shin,1 Mo K. Kang,1 Erica Dicterow,1 INTRODUCTION Ayako Kameta,1 Marcel A. Baluda,1 and Telomerase, which consists of the catalytic protein subunit, No-Hee Park1,2 human telomerase reverse transcriptase (hTERT), the RNA component of telomerase (hTR), and several associated pro- 1School of Dentistry and 2Jonsson Comprehensive Cancer Center, University of California, Los Angeles, California teins, has been primarily associated with maintaining the integ- rity of cellular DNA telomeres in normal cells (1, 2). Telomer- ase activity is correlated with the expression of hTERT, but not ABSTRACT with that of hTR (3, 4). Purpose: From numerous reports on proteins involved The involvement of DNA repair proteins in telomere main- in DNA repair and telomere maintenance that physically tenance has been well documented (5–8). In eukaryotic cells, associate with human telomerase reverse transcriptase nonhomologous end-joining requires a DNA ligase and the (hTERT), we inferred that hTERT/telomerase might play a DNA-activated protein kinase, which is recruited to the DNA role in DNA repair. We investigated this possibility in nor- ends by the DNA-binding protein Ku. Ku binds to hTERT mal human oral fibroblasts (NHOF) with and without ec- without the need for telomeric DNA or hTR (9), binds the topic expression of hTERT/telomerase. telomere repeat-binding proteins TRF1 (10) and TRF2 (11), and Experimental Design: To study the effect of hTERT/ is thought to regulate the access of telomerase to telomere DNA telomerase on DNA repair, we examined the mutation fre- ends (12, 13). -

(NAP) and Illustration of DNA Flexure Angles at Single Molecule Resolution Debayan Purkait†A, Debolina Bandyopadhyay†A, and Padmaja P
bioRxiv preprint doi: https://doi.org/10.1101/2020.09.11.293639; this version posted September 11, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Vital Insights into Prokaryotic Genome Compaction by Nucleoid-Associated Protein (NAP) and Illustration of DNA Flexure Angles at Single Molecule Resolution Debayan Purkait†a, Debolina Bandyopadhyay†a, and Padmaja P. Mishra*a aChemical Sciences Division, Saha Institute of Nuclear Physics, India aHomi Bhaba National Institute (HBNI), India †Both the authors have contributed equally. * Corresponding author Abstract Integration Host Factor (IHF) is a heterodimeric site-specific nucleoid-associated protein (NAP) well known for its DNA bending ability. The binding is mediated through the narrow minor grooves of the consensus sequence, involving van der-Waals interaction and hydrogen bonding. Although the DNA bend state of IHF has been captured by both X-ray Crystallography and Atomic Force Microscopy (AFM), the range of flexibility and degree of heterogeneity in terms of quantitative analysis of the nucleoprotein complex has largely remained unexplored. Here we have monitored and compared the trajectories of the conformational dynamics of a dsDNA upon binding of wild-type (wt) and single-chain (sc) IHF at millisecond resolution through single-molecule FRET (smFRET). Our findings reveal that the nucleoprotein complex exists in a ‘Slacked-Dynamic’ state throughout the observation window where many of them have switched between multiple ‘Wobbling States’ in the course of attainment of packaged form. A range of DNA ‘Flexure Angles’ has been calculated that give us vital insights regarding the nucleoid organization and transcriptional regulation in prokaryotes. -

Cell Cycle Regulation by the Bacterial Nucleoid
Available online at www.sciencedirect.com ScienceDirect Cell cycle regulation by the bacterial nucleoid David William Adams, Ling Juan Wu and Jeff Errington Division site selection presents a fundamental challenge to all terminate in the terminus region (Ter; 1808). Once organisms. Bacterial cells are small and the chromosome chromosome replication and segregation are complete (nucleoid) often fills most of the cell volume. Thus, in order to the cell is ready to divide. In Bacteria this normally occurs maximise fitness and avoid damaging the genetic material, cell by binary fission and in almost all species this is initiated division must be tightly co-ordinated with chromosome by the assembly of the tubulin homologue FtsZ into a replication and segregation. To achieve this, bacteria employ a ring-like structure (‘Z-ring’) at the nascent division site number of different mechanisms to regulate division site (Figure 1) [1]. The Z-ring then functions as a dynamic selection. One such mechanism, termed nucleoid occlusion, platform for assembly of the division machinery [2,3]. Its allows the nucleoid to protect itself by acting as a template for central role in division also allows FtsZ to serve as a nucleoid occlusion factors, which prevent Z-ring assembly over regulatory hub for the majority of regulatory proteins the DNA. These factors are sequence-specific DNA-binding identified to date [2,4]. Nevertheless, the precise ultra- proteins that exploit the precise organisation of the nucleoid, structure of the Z-ring and whether or not it plays a direct allowing them to act as both spatial and temporal regulators of role in force-generation during division remains contro- bacterial cell division. -

M1224-100 Thermostable Rnase H
BioVision 03/17 For research use only Thermostable RNAse H CATALOG NO.: M1224-100 10X THERMOSTABLE RNAse H REACTION BUFFER: 500 mM Tris-HCl, 1000 mM NaCl, AMOUNT: 500 U (100 µl) 100 mM MgCl2, pH 7.5 PRODUCT SOURCE: Recombinant E. coli REACTION CONDITIONS: Use 1X Thermostable RNAse H Reaction Buffer and incubate CONCENTRATION: 100 U/µl at a chosen temperature between 65°C and 95°C (enzyme half-life is 2 hours at 70°C and 30 minutes at 95°C). FORM: Liquid. Enzyme supplied with 10X Reaction Buffer COMPONENTS: Product Name Size Part. No. Thermostable RNAse H (5 U/μl) 100 µl M1224-100-1 10X Thermostable RNAse Reaction Buffer 500 µl M1224-100-2 DESCRIPTION: Thermostable RNAse H (Ribonuclease H) is an endoribonuclease that specifically hydrolyzes the phosphodiester bonds of RNA strands in RNA-DNA hybrids. Unlike E. coli RNAse H which is inactivated at temperatures above 55°C, Thermostable RNase H can withstand much higher temperatures. These higher temperatures allow for higher hybridization stringency for RNA-DNA heteroduplexes resulting in more specific hydrolysis of RNA. Thermostable RNase H has optimal activity above 65°C and is active up to 95°C making it useful for a broad range of applications. APPLICATIONS: 1. High-stringency hybrid selection and mapping of mRNA structure 2. Removal of mRNA prior to synthesis of second strand cDNA 3. Removal of the poly(A) sequences from mRNA in the presence of oligo (dT) 4. Directed cleavage of RNA RELATED PRODUCTS: STORAGE CONDITIONS: Store at -20°C. Avoid repeated freeze-thaw cycles of all E. -

Catalytic Domain of Plasmid Pad1 Relaxase Trax Defines a Group Of
Catalytic domain of plasmid pAD1 relaxase TraX defines a group of relaxases related to restriction endonucleases María Victoria Franciaa,1, Don B. Clewellb,c, Fernando de la Cruzd, and Gabriel Moncaliánd aServicio de Microbiología, Hospital Universitario Marqués de Valdecilla e Instituto de Formación e Investigación Marqués de Valdecilla, Santander 39008, Spain; bDepartment of Biologic and Materials Sciences, University of Michigan School of Dentistry, Ann Arbor, MI 48109; cDepartment of Microbiology and Immunology, University of Michigan Medical School, Ann Arbor, MI 48109; and dDepartamento de Biología Molecular e Instituto de Biomedicina y Biotecnología de Cantabria, Universidad de Cantabria–Consejo Superior de Investigaciones Científicas–Sociedad para el Desarrollo Regional de Cantabria, Santander 39011, Spain Edited by Roy Curtiss III, Arizona State University, Tempe, AZ, and approved July 9, 2013 (received for review May 30, 2013) Plasmid pAD1 is a 60-kb conjugative element commonly found in known relaxases show two characteristic sequence motifs, motif I clinical isolates of Enterococcus faecalis. The relaxase TraX and the containing the catalytic Tyr residue (which covalently attaches to primary origin of transfer oriT2 are located close to each other and the 5′ end of the cleaved DNA) and motif III with the His-triad have been shown to be essential for conjugation. The oriT2 site essential for relaxase activity (facilitating the cleavage reaction contains a large inverted repeat (where the nic site is located) by activation of the catalytic Tyr). This His-triad has been used as adjacent to a series of short direct repeats. TraX does not show a relaxase diagnostic signature (12). fi any of the typical relaxase sequence motifs but is the prototype of Plasmid pAD1 relaxase, TraX, was identi ed (9) and shown to oriT2 a unique family of relaxases (MOB ). -

Arthur Kornberg Discovered (The First) DNA Polymerase Four
Arthur Kornberg discovered (the first) DNA polymerase Using an “in vitro” system for DNA polymerase activity: 1. Grow E. coli 2. Break open cells 3. Prepare soluble extract 4. Fractionate extract to resolve different proteins from each other; repeat; repeat 5. Search for DNA polymerase activity using an biochemical assay: incorporate radioactive building blocks into DNA chains Four requirements of DNA-templated (DNA-dependent) DNA polymerases • single-stranded template • deoxyribonucleotides with 5’ triphosphate (dNTPs) • magnesium ions • annealed primer with 3’ OH Synthesis ONLY occurs in the 5’-3’ direction Fig 4-1 E. coli DNA polymerase I 5’-3’ polymerase activity Primer has a 3’-OH Incoming dNTP has a 5’ triphosphate Pyrophosphate (PP) is lost when dNMP adds to the chain E. coli DNA polymerase I: 3 separable enzyme activities in 3 protein domains 5’-3’ polymerase + 3’-5’ exonuclease = Klenow fragment N C 5’-3’ exonuclease Fig 4-3 E. coli DNA polymerase I 3’-5’ exonuclease Opposite polarity compared to polymerase: polymerase activity must stop to allow 3’-5’ exonuclease activity No dNTP can be re-made in reversed 3’-5’ direction: dNMP released by hydrolysis of phosphodiester backboneFig 4-4 Proof-reading (editing) of misincorporated 3’ dNMP by the 3’-5’ exonuclease Fidelity is accuracy of template-cognate dNTP selection. It depends on the polymerase active site structure and the balance of competing polymerase and exonuclease activities. A mismatch disfavors extension and favors the exonuclease.Fig 4-5 Superimposed structure of the Klenow fragment of DNA pol I with two different DNAs “Fingers” “Thumb” “Palm” red/orange helix: 3’ in red is elongating blue/cyan helix: 3’ in blue is getting edited Fig 4-6 E. -

Relaxation Complexes of Plasmids Colel and Cole2:Unique Site of The
Proc. Nat. Acad. Sci. USA Vol. 71, No. 10, pp. 3854-3857, October 1974 Relaxation Complexes of Plasmids ColEl and ColE2: Unique Site of the Nick in the Open Circular DNA of the Relaxed Complexes (Escherichia coli/supercoiled DNA/endonuclease/restriction enzyme/DNA replication) MICHAEL A. LOVETT, DONALD G. GUINEY, AND DONALD R. HELINSKI Department of Biology, University of California, San Diego, La Jolla, Calif. 92037 Communicated by Stanley L. Miller, June 26, 1974 ABSTRACT The product of the induced relaxation of volved in the production of a single-strand cleavage in the supercoiled DNA-protein relaxation complexes of colicino- initiation of replication and/or conjugal transfer of plasmid genic factors El (ColEl) and E2 (CoIE2) is an open circular DNA molecule with a strand-specific nick. Cleavage of the DNA. An involvement in such processes would most likely open circular DNA of each relaxed complex with the EcoRI require that the relaxation event take place at a unique site restriction endonuclease demonstrates that the single- in the DNA molecule. In this report, evidence will be pre- strand break is at a unique position. The site of the single- sented from studies using the EcoRI restriction endonuclease strand break in the relaxed ColEl complex is approximately the same distance from the EcoRI cleavage site as the that the strand-specific relaxation event takes place at a origin of ColEl DNA replication. unique site on both the ColEl and CoIE2 plasmid molecules. The location of this site, at least in the case of the ColEl Many of extrachromosomal circular DNA elements (plas- plasmid, is approximately the same distance from the single mids) of Escherichia coli, differing in molecular weight, num- EcoRI site as the origin of replication for this plasmid. -

The Response to DNA Damage at Telomeric Repeats and Its Consequences for Telomere Function
Review The Response to DNA Damage at Telomeric Repeats and Its Consequences for Telomere Function Ylli Doksani IFOM, The FIRC Institute of Molecular Oncology, via Adamello 16, 20139 Milan, Italy; [email protected]; Tel.: +39-02-574303258 Received: 26 March 2019; Accepted: 18 April 2019; Published: 24 April 2019 Abstract: Telomeric repeats, coated by the shelterin complex, prevent inappropriate activation of the DNA damage response at the ends of linear chromosomes. Shelterin has evolved distinct solutions to protect telomeres from different aspects of the DNA damage response. These solutions include formation of t-loops, which can sequester the chromosome terminus from DNA-end sensors and inhibition of key steps in the DNA damage response. While blocking the DNA damage response at chromosome ends, telomeres make wide use of many of its players to deal with exogenous damage and replication stress. This review focuses on the interplay between the end-protection functions and the response to DNA damage occurring inside the telomeric repeats, as well as on the consequences that telomere damage has on telomere structure and function. Keywords: telomere maintenance; shelterin complex; end-protection problem; telomere damage; telomeric double strand breaks; telomere replication; alternative lengthening of telomeres 1. Telomere Structure and End-Protection Functions Mammalian telomeres are made of tandem TTAGGG repeats that extend several kilobases and terminate with a 3′ single-stranded overhang about 50–400 nucleotides long. Telomeric repeats are coated in a sequence-specific fashion by a 6-protein complex named shelterin that prevents the activation of the Double Strand Break (DSB) response at chromosome ends [1-3]. -

T4 DNA Ligase Protocol
Certificate of Analysis T4 DNA Ligase: Part# 9PIM180 Size Part No. (Weiss units) M180A 100 Revised 4/18 M180B 500 M179A (High Conc.) 500 Ligase Buffer, 10X (C126A, C126B): The Ligase 10X Buffer supplied with this enzyme has a composition of 300mM Tris-HCl (pH 7.8), 100mM MgCl2, 100mM DTT and 10mM ATP. The performance of this buffer depends on the integrity of the ATP. Store the buffer in small aliquots at –20°C to minimize degradation of the ATP and DTT. *AF9PIM180 0418M180* Note: The DTT in the Ligase 10X Buffer may precipitate upon freezing. If this occurs, vortex the buffer until the precipitate AF9PIM180 0418M180 is in solution (typically 1–2 minutes). The performance of the product is not affected provided that the precipitate is resuspended. Enzyme Storage Buffer: T4 DNA Ligase is supplied in 10mM Tris-HCl (pH 7.4), 50mM KCl, 1mM DTT, 0.1mM EDTA and 50% glycerol. Source: E. coli strain expressing a recombinant clone. Unit Definition: 0.01 Weiss unit of T4 DNA Ligase is defined as the amount of enzyme required to catalyze the ligation of greater than 95% of the Hind III fragments of 1µg of Lambda DNA at 16°C in 20 minutes. See the unit concentration on the Product Information Label. Storage Temperature: Store at –20°C. Avoid multiple freeze-thaw cycles and exposure to frequent temperature changes. See the expiration date on the Product Information Label. Promega Corporation 2800 Woods Hollow Road Madison, WI 53711-5399 USA Quality Control Assays Telephone 608-274-4330 Toll Free 800-356-9526 Activity Assays Fax 608-277-2516 Internet www.promega.com Blue/White Assay: pGEM®-3Zf(+) Vector is digested with representative restriction enzymes (leaving 5´-termini, 3´-termini or blunt ends). -
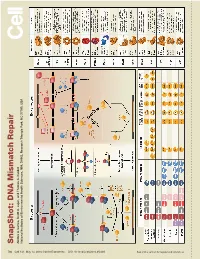
S Na P S H O T: D N a Mism a Tc H R E P a Ir
SnapShot: Repair DNA Mismatch Scott A. Lujan, and Thomas Kunkel A. Larrea, Andres Park, NC 27709, USA Triangle Health Sciences, NIH, DHHS, Research National Institutes of Environmental 730 Cell 141, May 14, 2010 ©2010 Elsevier Inc. DOI 10.1016/j.cell.2010.05.002 See online version for legend and references. SnapShot: DNA Mismatch Repair Andres A. Larrea, Scott A. Lujan, and Thomas A. Kunkel National Institutes of Environmental Health Sciences, NIH, DHHS, Research Triangle Park, NC 27709, USA Mismatch Repair in Bacteria and Eukaryotes Mismatch repair in the bacterium Escherichia coli is initiated when a homodimer of MutS binds as an asymmetric clamp to DNA containing a variety of base-base and insertion-deletion mismatches. The MutL homodimer then couples MutS recognition to the signal that distinguishes between the template and nascent DNA strands. In E. coli, the lack of adenine methylation, catalyzed by the DNA adenine methyltransferase (Dam) in newly synthesized GATC sequences, allows E. coli MutH to cleave the nascent strand. The resulting nick is used for mismatch removal involving the UvrD helicase, single-strand DNA-binding protein (SSB), and excision by single-stranded DNA exonucleases from either direction, depending upon the polarity of the nick relative to the mismatch. DNA polymerase III correctly resynthesizes DNA and ligase completes repair. In bacteria lacking Dam/MutH, as in eukaryotes, the signal for strand discrimination is uncertain but may be the DNA ends associated with replication forks. In these bacteria, MutL harbors a nick-dependent endonuclease that creates a nick that can be used for mismatch excision. Eukaryotic mismatch repair is similar, although it involves several dif- ferent MutS and MutL homologs: MutSα (MSH2/MSH6) recognizes single base-base mismatches and 1–2 base insertion/deletions; MutSβ (MSH2/MSH3) recognizes insertion/ deletion mismatches containing two or more extra bases. -

The Conjugative Relaxase Trwc Promotes Integration of Foreign
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by UCrea 1 The Conjugative Relaxase TrwC Promotes Integration of Foreign 2 DNA in the Human Genome 3 4 5 Coral González-Prieto,a Richard Gabriel,b Christoph Dehio,c Manfred Schmidt,b 6 and Matxalen Llosa,a# 7 8 Instituto de Biomedicina y Biotecnología de Cantabria (UC-CSIC-SODERCAN), 9 Santander, Spaina; Department of Translational Oncology, National Center for Tumor 10 Diseases and German Cancer Research Center, Heidelberg, Germanyb, Focal Area 11 Infection Biology, Biozentrum, Universität Basel, Basel, Switzerland 12 13 14 Running head: TrwC promotes DNA integration in the human genome 15 16 # Address correspondence to Matxalen LLosa, [email protected] 17 1 18 ABSTRACT 19 20 Bacterial conjugation is a mechanism of horizontal DNA transfer. The relaxase 21 TrwC of the conjugative plasmid R388 cleaves one strand of the transferred DNA at the 22 oriT, covalently attaches to it and leads the ssDNA into the recipient cell. In addition, 23 TrwC catalyzes site-specific integration of the transferred DNA into its target sequence 24 present in the genome of the recipient bacterium. Here, we report the analysis of the 25 efficiency and specificity of the integrase activity of TrwC in human cells, using the 26 Type IV Secretion System of the human pathogen Bartonella henselae to introduce 27 relaxase-DNA complexes. When compared to Mob relaxase from plasmid pBGR1, we 28 found that TrwC mediated a 10-fold increase in the rate of plasmid DNA transfer to 29 human cells, and a 100-fold increase in the rate of chromosomal integration of the 30 transferred DNA. -

Effects of Abasic Sites and DNA Single-Strand Breaks on Prokaryotic RNA Polymerases (Apurinic/Apyrimidinic Sites/Transcription/DNA Damage) WEI ZHOU* and PAUL W
Proc. Natl. Acad. Sci. USA Vol. 90, pp. 6601-6605, July 1993 Biochemistry Effects of abasic sites and DNA single-strand breaks on prokaryotic RNA polymerases (apurinic/apyrimidinic sites/transcription/DNA damage) WEI ZHOU* AND PAUL W. DOETSCH*tt Departments of *Biochemistry and tRadiation Oncology and tDivision of Cancer Biology, Emory University School of Medicine, Atlanta, GA 30322 Communicated by Philip C. Hanawalt, April 19, 1993 ABSTRACT Abasic sites are thought to be the most fre- effects of such damage on both transcription initiation and quently occurring cellular DNA damage and are generated elongation processes (19, 20). We wished to determine the spontaneously or as the result of chemical or radiation damage effects of AP sites on the transcription elongation process by to DNA. In contrast to the wealth of information that exists on using a defined system that would provide direct information the effects of abasic sites on DNA polymerases, very little is regarding the ability of RNA polymerases to bypass or be known about how these lesions interact with RNA polymerases. blocked by AP sites and, if bypass was observed, to deter- An in vitro transcription system was used to determine the mine the nature of the base insertion opposite such lesions. effects of abasic sites and single-strand breaks on transcrip- For these studies, we constructed a DNA template containing tional elongation. DNA templates were constructed containing a single AP site placed at a unique location downstream from single abasic sites or nicks placed at unique locations down- either the SP6 or tac promoter and carried out in vitro stream from two different promoters and were transcribed by transcription experiments with SP6 and E.