In Situ Microfluidic Cryofixation for Cryo Focused Ion Beam Milling and Cryo Electron Tomography
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Rapid Cryogenic Fixation of Biological Specimens for Electron Microscopy
RAPID CRYOGENIC FIXATION OF BIOLOGICAL SPECIMENS FOR ELECTRON MICROSCOPY KEITH PATRICKRYAN A thesis submitted in partial fulfilment of the requirements of the Council for National Academic Awards for the degree of Doctor of Philosophy September 1991 Polytechnic South West in collaboration with the Marine Biological Association of the United Kingdom and Plymouth Marine Laboratory ----- . \ ~ ,, '' - .... .._~ .. ·=·~-·-'-·-'" --······ --~....... ~=.sn.-.......... .r.=-..-> POL VTECHNIC SOUTH WEST liBRARY SERVICES Item C!O 00 7 9 4 9 3-0 No. ', )'; I Class 1 !) -, C!f -RYA !No. rr ... · ,Contl No. :x70ZS\0253 ·. ' COPYRIGHT This copy of the thesis has been supplied on condition that anyone who consults it is understood to recognise that its copyright rests with its author and that no quotation from the thesis and no information derived from it may be published without the authors prior written consent. 2 CONTENTS Page List of Figures 8 List of Tables 10 Abstract 11 Acknowledgements 12 1 Introduction 13 2 Literature Review 23 2.1 Background to specimen preservation for microscopy 23 2.2 Problems of chemical processing for electron rhlcroscopy 23 2.3 Introduction of cryotechniques into microscopy methods 24 2.4 The potential of cryofixation 25 2.5 The problems of cryofixation 25 2.6 Water, cooling and crystal nucleation 26 2. 7 Cell water 29 2.8 Crystallisation and latent heat release 29 2.9 Phase separation and eutectic temperature 30 2.10 Types of ice 31 2.11 Phase transitions 32 2.12 Ice crystal growth in frozen specimens after freezing 34 2.13 Cryoprotection against ice crystal damage 37 2.14 Modelling the cooling process 38 2.15 Cooling methods 45 2.16 Coolants (liquid) 46 2.17 Coolants (solid) 46 2.18 Plunge cooling methods 48 2.19 Jet cooling methods 51 2.20 Cryoblock methods 54 2.21 Rapid cooling experiments 59 3 2.22 Specimen rewarming during handling after freezing 74 2.23 Conclusions 74 3. -

Cryofixing Single Cells and Multicellular Specimens Enhances Structure and Immunocytochemistry for Light Microscopy TI Baskin
University of Massachusetts Amherst ScholarWorks@UMass Amherst Biology Department Faculty Publication Series Biology 1996 Cryofixing single cells and multicellular specimens enhances structure and immunocytochemistry for light microscopy TI Baskin DD Miller JW Vos JE Wilson PK Hepler Follow this and additional works at: https://scholarworks.umass.edu/biology_faculty_pubs Part of the Biology Commons Recommended Citation Baskin, TI; Miller, DD; Vos, JW; Wilson, JE; and Hepler, PK, "Cryofixing single cells and multicellular specimens enhances structure and immunocytochemistry for light microscopy" (1996). Journal of Microscopy-Oxford. 30. https://10.1046/j.1365-2818.1996.135417.x This Article is brought to you for free and open access by the Biology at ScholarWorks@UMass Amherst. It has been accepted for inclusion in Biology Department Faculty Publication Series by an authorized administrator of ScholarWorks@UMass Amherst. For more information, please contact [email protected]. Cryofixing single cells and multicellular specimens enhances structure and immunocytochemistry for light microscopy T. I. BASKIN,* D. D. MILLER,† J. W. VOS,‡ J. E. WILSON* & P. K. HEPLER†‡ *Division of Biological Sciences, University of Missouri, Columbia, MO 65211, U.S.A. †Molecular and Cellular Biology Program, and ‡Biology Department, University of Massachusetts, Amherst, MA 01003, U.S.A. Key words. Actin, chemical fixation, cryofixation, immunocytochemistry, methacrylate, microtubules, plunge freezing, pollen tubes, removable embedment, roots, stamen hairs. Summary crystals may be less severe than artefacts from chemical fixation. Cryofixation is widely held to be superior to chemical fixation for preserving cell structure; however, the use of cryofixation has been limited chiefly to electron microscopy. To see if Introduction cryofixation would improve sample structure or antigenicity It has long been realized that cryofixation is the method of as observed through the light microscope, we cryofixed choice for ultrastructural preservation of biological material. -

Aspects of Cryofixation and Cryosectioning for the Observation of Bulk Biological Samples in the Hydrated State by Cryoelectron Microscopy
Scanning Microscopy Volume 1996 Number 10 The Science of Biological Specimen Article 30 Preparation for Microscopy 12-17-1996 Aspects of Cryofixation and Cryosectioning for the Observation of Bulk Biological Samples in the Hydrated State by Cryoelectron Microscopy K. Richter University of Lausanne, Switzerland, [email protected] Follow this and additional works at: https://digitalcommons.usu.edu/microscopy Part of the Biology Commons Recommended Citation Richter, K. (1996) "Aspects of Cryofixation and Cryosectioning for the Observation of Bulk Biological Samples in the Hydrated State by Cryoelectron Microscopy," Scanning Microscopy: Vol. 1996 : No. 10 , Article 30. Available at: https://digitalcommons.usu.edu/microscopy/vol1996/iss10/30 This Article is brought to you for free and open access by the Western Dairy Center at DigitalCommons@USU. It has been accepted for inclusion in Scanning Microscopy by an authorized administrator of DigitalCommons@USU. For more information, please contact [email protected]. Scanning Microscopy Supplement 10, 1996 (pages 375-386) 0892-953X/96$5. 00 +. 25 Scanning Microscopy International, Chicago (AMF O'Hare), IL 60666 USA ASPECTS OF CRYOFIXATION AND CRYOSECTIONING FOR THE OBSERVATION OF BULK BIOLOGICAL SAMPLES IN THE HYDRATED STATE BY CRYOELECTRON MICROSCOPY K. Richter• University of Lausanne, Lausanne, Switzerland (Received for publication September 17, 1996 and in revised form December 17, 1996) Abstract Introduction Cryoelectron microscopy allows the observation of Biological specimens are not well suited to electron hydrated samples at high spatial resolution, and it would microscopy at room temperature. One major restriction be of great interest in biology to apply this method to is the vacuum in the microscope column, which does not cells and tissues. -

Microfluidic Cryofixation for Time-Correlated Live- Imaging, Cryo-Fluorescence Microscopy and Electron Microscopy of Caenorhabditis Elegans
Microfluidic cryofixation for time-correlated live- imaging, cryo-fluorescence microscopy and electron microscopy of Caenorhabditis elegans Dissertation for the award of the degree “Doctor rerum naturalium” of the Georg-August-Universität Göttingen within the doctoral program International Max Planck Research School “Physics of Biological and Complex Systems” of the Georg-August University School of Science (GAUSS) submitted by Giovanni Marco Nocera from Naples, Italy Göttingen, 2018 Thesis Committee Thomas Burg, PhD Max Planck Institute for Biophysical Chemistry, Göttingen Biological Micro- and Nanotechnology Prof. Dr. Sarah Köster Georg-August-University, Göttingen Institute for X-Ray Physics, Research Group Cellular Biophysics Prof. Dr. Stefan Hell Max Planck Institute for Biophysical Chemistry, Göttingen Dept. of NanoBiophotonics Members of the Examination Board Referee: Thomas Burg, PhD Max Planck Institute for Biophysical Chemistry, Göttingen Group of Biological Micro- and Nanotechnology 2nd Referee: Prof. Dr. Sarah Köster Georg-August-University, Göttingen Institute for X-Ray Physics, Group of Cellular Biophysics 3rd referee: Prof. Dr. Stefan Hell Max Planck Institute for Biophysical Chemistry, Göttingen Department of NanoBiophotonics Further members of the Examination Board Dr. Henrik Bringmann Max Planck Institute for Biophysical Chemistry, Göttingen Group of Sleep and Waking Prof. Dr. Silvio Rizzoli University Medical Center, Göttingen Department of Neuro- and Sensory Physiology Prof. Dr. Jörg Enderlein Georg-August-University, -

Electron Microscopy of High Pressure Frozen Samples: Bridging the Gap Between Cellular Ultrastructure and Atomic Resolution
Histochem Cell Biol (2008) 130:877–889 DOI 10.1007/s00418-008-0500-1 REVIEW Electron microscopy of high pressure frozen samples: bridging the gap between cellular ultrastructure and atomic resolution Daniel Studer · Bruno M. Humbel · Matthias Chiquet Accepted: 22 August 2008 / Published online: 16 September 2008 © Springer-Verlag 2008 Abstract Transmission electron microscopy has provided Moreover, recent Wndings will be discussed showing that most of what is known about the ultrastructural organiza- molecular models of proteins can be Wtted into depicted tion of tissues, cells, and organelles. Due to tremendous organellar ultrastructure of images of frozen hydrated sec- advances in crystallography and magnetic resonance imag- tions. High pressure freezing of tissue is the base which ing, almost any protein can now be modeled at atomic reso- may lead to precise models of macromolecular assemblies lution. To fully understand the workings of biological in situ, and thus to a better understanding of the function of “nanomachines” it is necessary to obtain images of intact complex cellular structures. macromolecular assemblies in situ. Although the resolution power of electron microscopes is on the atomic scale, in Keywords High pressure freezing · CryoWxation · biological samples artifacts introduced by aldehyde Wxa- Electron microscopy · Electron tomography · tion, dehydration and staining, but also section thickness Freeze substitution · Frozen hydrated sections · reduces it to some nanometers. CryoWxation by high pres- Immunolabeling sure freezing circumvents many of the artifacts since it allows vitrifying biological samples of about 200 m in thickness and immobilizes complex macromolecular Introduction assemblies in their native state in situ. To exploit the per- fect structural preservation of frozen hydrated sections, Transmission electron microscopy (TEM) has proven cru- sophisticated instruments are needed, e.g., high voltage cial to the advancement of modern cell biology. -

High-Quality Ultrastructural Preservation Using Cryofixation for 3D Electron Microscopy Of
bioRxiv preprint doi: https://doi.org/10.1101/261594; this version posted February 7, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 2 High-quality ultrastructural preservation using cryofixation for 3D electron microscopy of 3 genetically labeled tissues 4 5 6 Tin Ki Tsang1*, Eric A. Bushong2*, Daniela Boassa2, Junru Hu2, Benedetto Romoli3, Sebastien 7 Phan2, Davide Dulcis3, Chih-Ying Su1d and Mark H. Ellisman2,4d 8 9 10 1Neurobiology Section, Division of Biological Sciences, 11 University of California, San Diego, La Jolla, CA 92093, USA 12 13 2National Center for Microscopy and Imaging Research, Center for Research in Biological 14 Systems, University of California, San Diego, La Jolla, CA 92093, USA 15 16 3Department of Psychiatry, School of Medicine, University of California, San Diego, La Jolla, CA 17 92093, USA 18 19 4Department of Neurosciences, School of Medicine 20 University of California, San Diego, La Jolla, CA 92093, USA 21 22 23 24 * These authors contributed equally to this work. 25 dCorrespondence: [email protected] (M.H.E); [email protected] (C.Y.S) 1 bioRxiv preprint doi: https://doi.org/10.1101/261594; this version posted February 7, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. -

Cryofixation of Heart Tissue for X-Ray Microanalysis
Scanning Microscopy Volume 3 Number 4 Article 25 12-4-1989 Cryofixation of Heart Tissue for X-Ray Microanalysis Alice Warley St. Thomas's Hospital Campus Follow this and additional works at: https://digitalcommons.usu.edu/microscopy Part of the Biology Commons Recommended Citation Warley, Alice (1989) "Cryofixation of Heart Tissue for X-Ray Microanalysis," Scanning Microscopy: Vol. 3 : No. 4 , Article 25. Available at: https://digitalcommons.usu.edu/microscopy/vol3/iss4/25 This Article is brought to you for free and open access by the Western Dairy Center at DigitalCommons@USU. It has been accepted for inclusion in Scanning Microscopy by an authorized administrator of DigitalCommons@USU. For more information, please contact [email protected]. Scanning Microscopy, Vol. 3, No. 4, 1989 (Pages 1247-1252) 0891-7035/89$3.00+.00 Scanning Microscopy International, Chicago (AMF O'Hare), IL 60666 USA CRYOFIXATION OF HEARTTISSUE FOR X-RAY MICROANALYSIS Alice Warley Division of Biochemistry U.M.D.S. St. Thomas's Hospital Campus Lambeth Palace Road London SEl 7EH, U.K. (Received for publication April 11, 1989, and in revised form December 04, 1989) Abstract Introduction Cryofixation of tissues is necessary to be able to Changes in the concentrations of the inorganic study the concentrations of elements by X-ray micro elements Na, Mg, Kand Ca are thought to control the analysis. Simple dissection of heart tissue fragments functions of many different cells. Although much is of the size needed for optimum cryofixation by the con known about total elemental concentrations especially ventional methods of plunge or slam freezing leads to of normal tissues, more remains to be learned par the development of ischaemia in the tissue fragments ticularly about elemental concentrations in individual and a consequent redistribution of the diffusible cells, subcellular organelles, and whether alterations elements. -
An Automated Device for Cryofixation of Specimens of Electron Microscopy Using Liquid Helium
Plant Cell Physiol. 42(9): 885–893 (2001) JSPP © 2001 Technical Advance: An Automated Device for Cryofixation of Specimens of Electron Microscopy using Liquid Helium Akiko Hisada 1, 4,TomokoYoshida1, 2, Shigeo Kubota 1, 5,NaokoK.Nishizawa3 and Masaki Furuya 1 1 Hitachi Advanced Research Laboratory, Hatoyama, Saitama, 350-0395 Japan 2 Hitachi Instruments Service Co., Yotsuya, Shinjuku-ku, Tokyo, 160-0004 Japan 3 The University of Tokyo, Yayoi, Bunkyo-ku, Tokyo, 113-8657 Japan ; Metal-contact rapid freezing using liquid helium is the- (TEM). For rapid processes, temporal resolution is also impor- oretically the best method for preserving the fine structure tant (Van Harreveld et al. 1974, Heuser et al. 1976, Heuser et of living cells with high temporal resolution in preparation al. 1979). However, TEM often fails to preserve the precise of tissue samples for electron microscopy. However, this structural information about dynamic cellular processes, method is not commonly used, because of its technical diffi- because conventional processing consists of slow processes culty and low reproducibility. We have designed and con- such as chemical fixation and dehydration at room tempera- structed an automatic device which allows simple, rapid ture. The limitations of chemical fixation can largely be over- and reproducible preparation of high-quality electron come using the cryofixation technique. The advantage of cry- microscopic specimens by the non-specialist. We assessed ofixation over conventional processing lies mainly in the the quality of cryofixation in samples prepared using this extremely rapid physical fixation of the living specimen. The device by examining the preservation of cellular ultrastruc- time required for cryofixation by vitrification (absence of crys- ture in relation to distance from the freezing block, and talline ice in a specimen) is estimated to be less than 0.1 ms, found that the region within 10 mmofthemetal-contact which is substantially less (by a factor of 104) than that plane was fixed with the highest quality. -
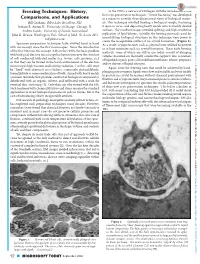
Freezing Techniques: History, in the 1970’S, a New Era of EM Began with the Introduction of the First Cryo-Preservation Techniques
Freezing Techniques: History, In the 1970’s, a new era of EM began with the introduction of the first cryo-preservation techniques. “Freeze fracturing” was developed Downloaded from Comparisons, and Applications as a means to provide three-dimensional views of biological materi- Bill Graham, Bibst Labs Brookline, NH als. This technique involved freezing a biological sample, fracturing Jotham R. Austin II, University of Chicago Chicago, IL it open in vacuo, and depositing heavy metals onto its freshly cleaved surfaces. The resultant images revealed splitting and high-resolution Andres Kaech, University of Zurich, Switzerland https://www.cambridge.org/core John E. Heuser, Washington Univ. School of Med., St. Louis, MO replication of lipid bilayers. Initially, the freezing protocols used for immobilizing biological structures in this technique were prone to [email protected] create the recognizable artifact of ice crystal formation. (Figure 1). Specimen preparation techniques have evolved hand in hand As a result, cryoprotectants such as glycerol were utilized to prevent with microscopy since the first microscopes. Since the introduction or at least minimize such ice crystal formation. These early freezing of the first Electron Microscope (EM) in the 1930’s, the basic problem methods, some of which are still in use today, consist of plunging with biological electron microscopy has been to preserve the structure samples mounted on thermally conductive supports into containers of soft condensed, hydrated matter (e.g. tissues, cells, proteins, etc.) of liquefied organic gases (chlorodifluoromethanes, ethane, propane), so that they can be viewed in the harsh environment of the electron and/or slurries of liquid nitrogen. -

Practical Methods in High- Pressure Freezing, Freeze-Substitution, Embedding and Immunocytochemistry for Electron Microscopy
Practical Methods in High- Pressure Freezing, Freeze-Substitution, Embedding and Immunocytochemistry for Electron Microscopy Mary K Morphew Laboratory for 3-D Fine Structure Dept. of MCD Biology University of Colorado Boulder, Colorado 1 Table of Contents Introduction Chapter 1. Rapid Freezing by High Pressure A) Theory B) Preparation C) Specimen Holders D) Additives E) Sample Loading 1) Suspensions 2) Monolayers 3) Tissues 4) Embryos Chapter 2. Freeze Substitution A) Equipment B) Recipes Chapter 3. Embedding and Sectioning A) Ultrastructure B) Immunocytochemistry Chapter 4. Immunocytochemistry A) Sectioning B) Immunostaining Preparation C) Antibody Applications References Appendix 1: Useful Articles Appendix 2: Vendor Information 2 Practical Methods in High Pressure Rapid Freezing, Freeze Substitution, Embedding and Immunocytochemistry for Electron Microscopy This manual is a technical guide to methods in high pressure freezing and subsequent freeze-substitution fixation and embedding for high resolution electron microscopy and EM- immunocytochemistry. It contains a number of methodologies which we have found useful in various combinations to provide acceptable-to-superior preservation of a variety of biological samples. Many of these methodologies are relatively new and still require experimentation for certain biological systems; however, this manual will provide some critical ground work for future investigations. 3 Introduction Fixation of biological samples for preservation of fine structure has been a goal of structural biologists since the beginnings of electron microscopy. The goal has always been to preserve samples as close to their native state as possible with resolution that could be achieved by the technology of their time. As microscopic technology and computer capabilities have grown so has interest in the rapid fixation of biological samples.