Comparative Genomic Analysis of Arctic Permafrost Bacterium Nesterenkonia Sp
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Analysis of the Impact of Silver Ions on Creatine Amidinohydrolase
ActaBIOMATERIALIA Acta Biomaterialia 1 (2005) 183–191 www.actamat-journals.com A stable three enzyme creatinine biosensor. 2. Analysis of the impact of silver ions on creatine amidinohydrolase Jason A. Berberich b,1, Lee Wei Yang a, Ivet Bahar a, Alan J. Russell b,* a Center for Computational Biology & Bioinformatics and Department of Molecular Genetics & Biochemistry, School of Medicine, University of Pittsburgh, Pittsburgh, PA, USA b Department of Surgery, McGowan Institute for Regenerative Medicine, University of Pittsburgh, Pittsburgh, PA 15219, USA Received 11 October 2004; received in revised form 26 November 2004; accepted 28 November 2004 Abstract The enzyme creatine amidinohydrolase is a clinically important enzyme used in the determination of creatinine in blood and urine. Continuous use biosensors are becoming more important in the clinical setting; however, long-use creatinine biosensors have not been commercialized due to the complexity of the three-enzyme creatinine biosensor and the lack of stability of its components. This paper, the second in a series of three, describes the immobilization and stabilization of creatine amidinohydrolase. Creatine amidinohydrolase modified with poly(ethylene glycol) activated with isocyanate retains significant activity after modification. The enzyme was successfully immobilized into hydrophilic polyurethanes using a reactive prepolymer strategy. The immobilized enzyme retained significant activity over a 30 day period at 37 °C and was irreversibly immobilized into the polymer. Despite being stabilized in the polymer, the enzyme remained highly sensitive to silver ions which were released from the amperometric electrodes. Computational analysis of the structure of the protein using the Gaussian network model suggests that the silver ions bind tightly to a cysteine residue preventing normal enzyme dynamics and catalysis. -

Reduction of Pectinesterase Activity in a Commercial Enzyme Preparation
Journal of the Science of Food and Agriculture J Sci Food Agric 85:1613–1621 (2005) DOI: 10.1002/jsfa.2154 Reduction of pectinesterase activity in a commercial enzyme preparation by pulsed electric fields: comparison of inactivation kinetic models Joaquın´ Giner, Pascal Grouberman, Vicente Gimeno and Olga Martın´ ∗ Department of Food Technology, University of Lleida, CeRTA-UTPV, ETSEA, Avda Alcalde Rovira Roure 191, 25198-Lleida, Spain Abstract: The inactivation of pectinesterase (PE) in a commercial enzyme preparation (CEP) under high intensity pulsed electric fields (HIPEF) was studied. After desalting and water dilution of the raw CEP, samples were exposed to exponentially decay waveform pulses for up to 463 µs at electric field intensities ranging from 19 to 38 kV cm−1. Pulses were applied in monopolar mode. Experimental data were fitted to a first-order kinetic model as well as to models based on Fermi, Hulsheger¨ or Weibull equations to describe PE inactivation kinetics. Characteristic parameters for each model were calculated. Relationships between some of the parameters and process variables were obtained. The Weibull model yielded the best accuracy factor. The relationship between residual PE and input of electrical energy density was found to be that of exponential decay. 2005 Society of Chemical Industry Keywords: pulsed electric fields; kinetics; pectinesterase; model; inactivation INTRODUCTION It has become customary to use CEPs in fruit and Pectinesterase (PE; EC 3.1.1.11) is a pectic enzyme vegetable juice technology. Depending -

Fruit Ripening and Storage
OPEN Citation: Horticulture Research (2014) 1, 6; doi:10.1038/hortres.2014.6 ß 2014 Nanjing Agricultural University All rights reserved 2052-7276/14 www.nature.com/hortres ARTICLE Dynamic changes in proteins during apple (Malus x domestica) fruit ripening and storage Yun Shi1, Li Jiang1, Li Zhang2, Ruoyi Kang1 and Zhifang Yu1 A proteomic study, using two-dimensional polyacrylamide gel electrophoresis and matrix-assisted laser desorption/ionization time-of-flight/time-of-flight, was conducted in apple fruit (cv. ‘Golden Delicious’) starting at 10 days prior to harvest through 50 days in storage. Total protein was extracted using a phenol/sodium dodecyl sulfate protocol. More than 400 protein spots were detected in each gel and 55 differentially expressed proteins (p,0.05) were subjected to matrix-assisted laser desorption/ionization time-of-flight/ time-of-flight analysis. Fifty-three of these proteins were finally identified using an apple expressed sequence tag database downloaded from Genome Database for Rosaceae and placed into six categories. The categories and the percentage of proteins placed in each category were stress response and defense (49.0%), energy and metabolism (34.0%), fruit ripening and senescence (5.6%), signal transduction (3.8%), cell structure (3.8%) and protein synthesis (3.8%). Proteins involved in several multiple metabolic pathways, including glycolysis, pentose–phosphate pathway, anti-oxidative systems, photosynthesis and cell wall synthesis, were downregulated, especially during the climacteric burst in respiration and during the senescent stages of fruit development. Proteins classified as allergens or involved in cell wall degradation were upregulated during the ripening process. Some protein spots exhibited a mixed pattern (increasing to maximal abundance followed by a decrease), such as 1-aminocyclopropane-1-carboxylate oxidase, L-ascorbate peroxidase and abscisic acid response proteins. -

Genome-Wide Analysis of Glyoxalase-Like Gene Families in Grape
Li et al. BMC Genomics (2019) 20:362 https://doi.org/10.1186/s12864-019-5733-y RESEARCHARTICLE Open Access Genome-wide analysis of glyoxalase-like gene families in grape (Vitis vinifera L.) and their expression profiling in response to downy mildew infection Tiemei Li1,2,3, Xin Cheng1,2,3, Yuting Wang1,2,3, Xiao Yin1,2,3, Zhiqian Li1,2,3, Ruiqi Liu1,2,3, Guotian Liu1,2,3, Yuejin Wang1,2,3 and Yan Xu1,2,3* Abstract Background: The glyoxalase system usually comprises two enzymes, glyoxalase I (GLYI) and glyoxalase II (GLYII). This system converts cytotoxic methylglyoxal (MG) into non-toxic D-lactate in the presence of reduced glutathione (GSH) in two enzymatic steps. Recently, a novel type of glyoxalase III (GLYIII) activity has observed in Escherichia coli that can detoxify MG into D-lactate directly, in one step, without a cofactor. Investigation of the glyoxalase enzymes of a number of plant species shows the importance of their roles in response both to abiotic and to biotic stresses. Until now, glyoxalase gene families have been identified in the genomes of four plants, Arabidopsis, Oryza sativa, Glycine max and Medicago truncatula but no similar study has been done with the grapevine Vitis vinifera L. Results: In this study, four GLYI-like,twoGLYII-like and three GLYIII-like genesareidentifiedfromthegenomedatabaseof grape. All these genes were analysed in detail, including their chromosomal locations, phylogenetic relationships, exon-intron distributions, protein domain organisations and the presence of conserved binding sites. Using quantitative real-time PCR analysis (qRT-PCR), the expression profiles of these geneswereanalysedindifferent tissues of grape, and also when under infection stress from downy mildew (Plasmopara viticola). -

Phytic Acid (Phytate)/ Total Phosphorus
www.megazyme.com PHYTIC ACID (PHYTATE)/ TOTAL PHOSPHORUS Measured as phosphorus released by phytase and alkaline phosphatase ASSAY PROCEDURE K-PHYT 05/19 (50 Assays per Kit) © Megazyme 2019 INTRODUCTION: Phytic acid (phytate; myo-inositol 1,2,3,4,5,6-hexakisphosphate) is the primary source of inositol and storage phosphorus in plant seeds contributing ~ 70% of total phosphorus. The abundance of phytic acid in cereal grains is a concern in the foods and animal feeds industries because the phosphorus in this form is unavailable to monogastric animals due to a lack of endogenous phytases; enzymes specific for the dephosphorylation of phytic acid. In addition, the strong chelating characteristic of phytic acid reduces the bioavailability of other essential dietary nutrients such as minerals (e.g. Ca2+, Zn2+, Mg2+, Mn2+, Fe2+/3+), proteins and amino acids.2 High phytic acid content feeds are generally supplemented with inorganic phosphate, however this causes increased faecal phosphate levels and subsequent eutrophication of waterways. Alternatively, supplementation with commercial phytases is becoming increasingly popular and reduces the requirement for inorganic phosphate supplementation as well as the associated environmental issues. Currently, there is no commercially available, simple, quantitative method for phytic acid and, while such measurement is relatively complex, the generally accepted AOAC Method 986.11 has limitations.3 For each individual analysis the method requires cumbersome anion-exchange purification and a major inherent assumption here is that only phytic acid is purified. While this assumption is viable for non-processed grains for which phytic acid comprises at least 97% of total inositol phosphates, it is not viable for processed foods and feeds which can contain higher levels of some lower myo-inositol phosphate forms (i.e. -

Effect of Ph and Temperature on the Activity of Phytase Products Used In
Brazilian Journal of Poultry Science Revista Brasileira de Ciência Avícola Effect of ph and Temperature on the Activity of ISSN 1516-635X Jul - Sept 2012/ v.14 / n.3 / 159-232 Phytase Products Used in Broiler Nutrition Author(s) ABSTRACT Naves L de P1 Corrêa AD2 The activity of three commercial microbial phytase (Aspergillus Bertechini AG3 oryzae, A. niger, and Saccharomyces cerevisae) products used in broiler Gomide EM4 Santos CD dos2 nutrition was determined at different pH (2.0 to 9.0) and temperature (20 to 90°C) values. Enzymatic activity was determined according to the reaction of the phytase with its substrate (sodium phytate), in four replicates, and was expressed in units of phytase activity (FTU). A. oryzae phytase exhibited optimal activity at pH 4.0 and 40°C, but 1Graduate student in Monogastric Nutrition of its absolute activity was the lowest of the three phytases evaluated. the Animal Science Department − Federal A. niger phytase exhibited maximal activity close to pH 5.0 and 45oC, University of Lavras (UFLA). whereas S. cerevisae phytase presented its highest activity at pH close to 2Professor of the Chemistry Department/ UFLA. 4.5 and temperatures ranging between 50 and 60°C. It was concluded 3Professor of the Animal Science Department/ that A. niger and S. cerevisae phytase products exhibited the highest UFLA. absolute activities in vitro at pH and temperature values (pH lower than 4Ph. D. student in Monogastric Nutrition of o the Animal Science Department/UFLA. 5.0 and 41 C) corresponding to the ideal physiological conditions of broilers, which would theoretically allow high hydrolysis rate of the phytate contained in the feed. -

Peraturan Badan Pengawas Obat Dan Makanan Nomor 28 Tahun 2019 Tentang Bahan Penolong Dalam Pengolahan Pangan
BADAN PENGAWAS OBAT DAN MAKANAN REPUBLIK INDONESIA PERATURAN BADAN PENGAWAS OBAT DAN MAKANAN NOMOR 28 TAHUN 2019 TENTANG BAHAN PENOLONG DALAM PENGOLAHAN PANGAN DENGAN RAHMAT TUHAN YANG MAHA ESA KEPALA BADAN PENGAWAS OBAT DAN MAKANAN, Menimbang : a. bahwa masyarakat perlu dilindungi dari penggunaan bahan penolong yang tidak memenuhi persyaratan kesehatan; b. bahwa pengaturan terhadap Bahan Penolong dalam Peraturan Kepala Badan Pengawas Obat dan Makanan Nomor 10 Tahun 2016 tentang Penggunaan Bahan Penolong Golongan Enzim dan Golongan Penjerap Enzim dalam Pengolahan Pangan dan Peraturan Kepala Badan Pengawas Obat dan Makanan Nomor 7 Tahun 2015 tentang Penggunaan Amonium Sulfat sebagai Bahan Penolong dalam Proses Pengolahan Nata de Coco sudah tidak sesuai dengan kebutuhan hukum serta perkembangan ilmu pengetahuan dan teknologi sehingga perlu diganti; c. bahwa berdasarkan pertimbangan sebagaimana dimaksud dalam huruf a dan huruf b, perlu menetapkan Peraturan Badan Pengawas Obat dan Makanan tentang Bahan Penolong dalam Pengolahan Pangan; -2- Mengingat : 1. Undang-Undang Nomor 18 Tahun 2012 tentang Pangan (Lembaran Negara Republik Indonesia Tahun 2012 Nomor 227, Tambahan Lembaran Negara Republik Indonesia Nomor 5360); 2. Peraturan Pemerintah Nomor 28 Tahun 2004 tentang Keamanan, Mutu dan Gizi Pangan (Lembaran Negara Republik Indonesia Tahun 2004 Nomor 107, Tambahan Lembaran Negara Republik Indonesia Nomor 4424); 3. Peraturan Presiden Nomor 80 Tahun 2017 tentang Badan Pengawas Obat dan Makanan (Lembaran Negara Republik Indonesia Tahun 2017 Nomor 180); 4. Peraturan Badan Pengawas Obat dan Makanan Nomor 12 Tahun 2018 tentang Organisasi dan Tata Kerja Unit Pelaksana Teknis di Lingkungan Badan Pengawas Obat dan Makanan (Berita Negara Republik Indonesia Tahun 2018 Nomor 784); MEMUTUSKAN: Menetapkan : PERATURAN BADAN PENGAWAS OBAT DAN MAKANAN TENTANG BAHAN PENOLONG DALAM PENGOLAHAN PANGAN. -

Download Product Insert (PDF)
PRODUCT INFORMATION Lysophospholipase D Polyclonal Antibody Item No. 10005375 Overview and Properties Contents: This vial contains 500 µl of peptide affinity-purified antibody. Synonyms: Autotaxin, ENPP2, lysoPLD Immunogen: Peptide from the C-terminal region of rat LysoPLD Species Reactivity: (+) Human, mouse, and rat; other species not tested Form: Liquid Storage: -20°C (as supplied) Stability: ≥1 year Storage Buffer: TBS, pH 7.4, with 50% glycerol, 0.1%l BSA, and 0.02% sodium azide Host: Rabbit Applications: Immunocytochemistry (ICC), Immunohistochemistry (IHC), and Western blot (WB); the recommended starting dilution for ICC is 1:500, 1:80 for IHC, and 1:200 for WB. Other applications were not tested, therefore optimal working concentration/dilution should be determined empirically. Images 1 · · · · · · · 104 kDa · · · · · · · 60 kDa Lane 1: Human cerebella supernatant (40 µg) Immunohistochemistry analysis of formalin-fixed, paraffin-embedded (FFPE) human cerebellum ǎssue aer heat-induced anǎgen retrieval in pH 6.0 citrate buffer. Aer incubaǎon with Lysophospholipase D Polyclonal Anǎbody (Item No. 10005375) at a 1:80 diluǎon, slides were incubated with bioǎnylated secondary anǎbody, followed by alkaline phosphatase-strepavidin and chromogen (DAB). WARNING CAYMAN CHEMICAL THIS PRODUCT IS FOR RESEARCH ONLY - NOT FOR HUMAN OR VETERINARY DIAGNOSTIC OR THERAPEUTIC USE. 1180 EAST ELLSWORTH RD SAFETY DATA ANN ARBOR, MI 48108 · USA This material should be considered hazardous until further information becomes available. Do not ingest, inhale, get in eyes, on skin, or on clothing. Wash thoroughly after handling. Before use, the user must review the complete Safety Data Sheet, which has been sent via email to your institution. PHONE: [800] 364-9897 WARRANTY AND LIMITATION OF REMEDY [734] 971-3335 Buyer agrees to purchase the material subject to Cayman’s Terms and Conditions. -

Supplementary Materials
Supplementary Materials COMPARATIVE ANALYSIS OF THE TRANSCRIPTOME, PROTEOME AND miRNA PROFILE OF KUPFFER CELLS AND MONOCYTES Andrey Elchaninov1,3*, Anastasiya Lokhonina1,3, Maria Nikitina2, Polina Vishnyakova1,3, Andrey Makarov1, Irina Arutyunyan1, Anastasiya Poltavets1, Evgeniya Kananykhina2, Sergey Kovalchuk4, Evgeny Karpulevich5,6, Galina Bolshakova2, Gennady Sukhikh1, Timur Fatkhudinov2,3 1 Laboratory of Regenerative Medicine, National Medical Research Center for Obstetrics, Gynecology and Perinatology Named after Academician V.I. Kulakov of Ministry of Healthcare of Russian Federation, Moscow, Russia 2 Laboratory of Growth and Development, Scientific Research Institute of Human Morphology, Moscow, Russia 3 Histology Department, Medical Institute, Peoples' Friendship University of Russia, Moscow, Russia 4 Laboratory of Bioinformatic methods for Combinatorial Chemistry and Biology, Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry of the Russian Academy of Sciences, Moscow, Russia 5 Information Systems Department, Ivannikov Institute for System Programming of the Russian Academy of Sciences, Moscow, Russia 6 Genome Engineering Laboratory, Moscow Institute of Physics and Technology, Dolgoprudny, Moscow Region, Russia Figure S1. Flow cytometry analysis of unsorted blood sample. Representative forward, side scattering and histogram are shown. The proportions of negative cells were determined in relation to the isotype controls. The percentages of positive cells are indicated. The blue curve corresponds to the isotype control. Figure S2. Flow cytometry analysis of unsorted liver stromal cells. Representative forward, side scattering and histogram are shown. The proportions of negative cells were determined in relation to the isotype controls. The percentages of positive cells are indicated. The blue curve corresponds to the isotype control. Figure S3. MiRNAs expression analysis in monocytes and Kupffer cells. Full-length of heatmaps are presented. -
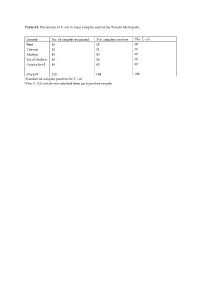
Supplementary File 1
Table S1. Prevalence of E. coli in meat samples sold at the Tamale Metropolis. Sample No. of samples examined aNo. samples positive bNo. E. coli Beef 45 39 39 Chevon 45 34 34 Mutton 45 40 40 Local chicken 45 36 36 Guinea fowl 45 40 40 Overall 225 189 189 aNumber of samples positive for E. coli. bOne E. Coli isolate was selected from each positive sample. Table S2. A table showing the eBURST (Based Upon Related Sequence Types) analyses of the study sequence types with global curated STs in Escherichia PubMLST database. MLST (Isolate) Type of clone Closet global ancestry Source sequence type (ST) ST69 (SG6) Similar a ST69 Animal (Food), Human ST155 (SLC2, Similar ST155 Animal (Food), Human, TLC13, CM4) Environment ST297 (TLC1) Similar ST297 Human ST1727 (NC3) Similar ST1727 Human ST44 (AC1) Single-Locus Variant ST10, ST752 Animal (Food), (SLV) b Human ST469 (CC6) Single-Locus Variant ST162 Food (SLV) ST540 (AB1, Single-Locus Variant ST4093 Human TG1) (SLV) ST1141 (NM11) Single-Locus Variant ST10, ST744 Animal (Food), (SLV) Human ST7473 (NB12) Single-Locus Variant ST10 Animal (Food), (SLV) Human ST6646 (CB1) Satellite c None - ST7483 (NB12) Satellite None - a Similar: study isolate was similar to a global curated known sequence type. b Single-Locus Variant (SLV): study isolate only shared similarity with global curated known sequence types that differed in one allelic gene. c Satellite: study isolate as a distantly related and did not shared any similarity with global curated known sequence types. Table S3. In silico identification and characterization of conserved stress response mechanisms in the E. -

Differential Gene Expression in Tomato Fruit and Colletotrichum
Barad et al. BMC Genomics (2017) 18:579 DOI 10.1186/s12864-017-3961-6 RESEARCH Open Access Differential gene expression in tomato fruit and Colletotrichum gloeosporioides during colonization of the RNAi–SlPH tomato line with reduced fruit acidity and higher pH Shiri Barad1,2, Noa Sela3, Amit K. Dubey1, Dilip Kumar1, Neta Luria1, Dana Ment1, Shahar Cohen4, Arthur A. Schaffer4 and Dov Prusky1* Abstract Background: The destructive phytopathogen Colletotrichum gloeosporioides causes anthracnose disease in fruit. During host colonization, it secretes ammonia, which modulates environmental pH and regulates gene expression, contributing to pathogenicity. However, the effect of host pH environment on pathogen colonization has never been evaluated. Development of an isogenic tomato line with reduced expression of the gene for acidity, SlPH (Solyc10g074790.1.1), enabled this analysis. Total RNA from C. gloeosporioides colonizing wild-type (WT) and RNAi– SlPH tomato lines was sequenced and gene-expression patterns were compared. Results: C. gloeosporioides inoculation of the RNAi–SlPH line with pH 5.96 compared to the WT line with pH 4.2 showed 30% higher colonization and reduced ammonia accumulation. Large-scale comparative transcriptome analysis of the colonized RNAi–SlPH and WT lines revealed their different mechanisms of colonization-pattern activation: whereas the WT tomato upregulated 13-LOX (lipoxygenase), jasmonic acid and glutamate biosynthesis pathways, it downregulated processes related to chlorogenic acid biosynthesis II, phenylpropanoid biosynthesis and hydroxycinnamic acid tyramine amide biosynthesis; the RNAi–SlPH line upregulated UDP-D-galacturonate biosynthesis I and free phenylpropanoid acid biosynthesis, but mainly downregulated pathways related to sugar metabolism, such as the glyoxylate cycle and L-arabinose degradation II. -

Final Dilution in Petri Plates with the Experiment, the Birds Were Weighed on Day-Of-Hatch and D 7, 10, Proper Culture Media, Tryptic Soy Agar (Biokar)
Symposium on Gut Health in Production of Food Animals November 10–12, 2014, St. Louis, Missouri Program and Abstracts www.GutHealthSymposium.com/2014 SYMPOSIUM ON GUT HEALTH IN PRODUCTION OF FOOD ANIMALS CONTENTS Welcome ............................................................................................................................3 Map ....................................................................................................................................4 Program .............................................................................................................................5 Poster Presentations......................................................................................................10 Abstracts .........................................................................................................................12 Poster Abstracts .............................................................................................................26 1 SYMPOSIUM ON GUT HEALTH IN PRODUCTION OF FOOD ANIMALS WELCOME On behalf of the Organizing Committee for the 3rd Symposium on Gut Health in Production of Food Animals, I welcome you to St. Louis, Missouri! I hope that the change in venue has made travel to the symposium easier and less expensive. Like the first two symposia organized around the topic of gut health in food animals, the aim this year is to bring together a group of scientists from academia, government, and industry to discuss the role of gut health in animal production and the essential