Dectin-1 Controls TLR9 Trafficking to Phagosomes Containing Β-1,3 Glucan Nida S
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse
Welcome to More Choice CD Marker Handbook For more information, please visit: Human bdbiosciences.com/eu/go/humancdmarkers Mouse bdbiosciences.com/eu/go/mousecdmarkers Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse CD3 CD3 CD (cluster of differentiation) molecules are cell surface markers T Cell CD4 CD4 useful for the identification and characterization of leukocytes. The CD CD8 CD8 nomenclature was developed and is maintained through the HLDA (Human Leukocyte Differentiation Antigens) workshop started in 1982. CD45R/B220 CD19 CD19 The goal is to provide standardization of monoclonal antibodies to B Cell CD20 CD22 (B cell activation marker) human antigens across laboratories. To characterize or “workshop” the antibodies, multiple laboratories carry out blind analyses of antibodies. These results independently validate antibody specificity. CD11c CD11c Dendritic Cell CD123 CD123 While the CD nomenclature has been developed for use with human antigens, it is applied to corresponding mouse antigens as well as antigens from other species. However, the mouse and other species NK Cell CD56 CD335 (NKp46) antibodies are not tested by HLDA. Human CD markers were reviewed by the HLDA. New CD markers Stem Cell/ CD34 CD34 were established at the HLDA9 meeting held in Barcelona in 2010. For Precursor hematopoetic stem cell only hematopoetic stem cell only additional information and CD markers please visit www.hcdm.org. Macrophage/ CD14 CD11b/ Mac-1 Monocyte CD33 Ly-71 (F4/80) CD66b Granulocyte CD66b Gr-1/Ly6G Ly6C CD41 CD41 CD61 (Integrin b3) CD61 Platelet CD9 CD62 CD62P (activated platelets) CD235a CD235a Erythrocyte Ter-119 CD146 MECA-32 CD106 CD146 Endothelial Cell CD31 CD62E (activated endothelial cells) Epithelial Cell CD236 CD326 (EPCAM1) For Research Use Only. -

Supplemental Figure 1. Vimentin
Double mutant specific genes Transcript gene_assignment Gene Symbol RefSeq FDR Fold- FDR Fold- FDR Fold- ID (single vs. Change (double Change (double Change wt) (single vs. wt) (double vs. single) (double vs. wt) vs. wt) vs. single) 10485013 BC085239 // 1110051M20Rik // RIKEN cDNA 1110051M20 gene // 2 E1 // 228356 /// NM 1110051M20Ri BC085239 0.164013 -1.38517 0.0345128 -2.24228 0.154535 -1.61877 k 10358717 NM_197990 // 1700025G04Rik // RIKEN cDNA 1700025G04 gene // 1 G2 // 69399 /// BC 1700025G04Rik NM_197990 0.142593 -1.37878 0.0212926 -3.13385 0.093068 -2.27291 10358713 NM_197990 // 1700025G04Rik // RIKEN cDNA 1700025G04 gene // 1 G2 // 69399 1700025G04Rik NM_197990 0.0655213 -1.71563 0.0222468 -2.32498 0.166843 -1.35517 10481312 NM_027283 // 1700026L06Rik // RIKEN cDNA 1700026L06 gene // 2 A3 // 69987 /// EN 1700026L06Rik NM_027283 0.0503754 -1.46385 0.0140999 -2.19537 0.0825609 -1.49972 10351465 BC150846 // 1700084C01Rik // RIKEN cDNA 1700084C01 gene // 1 H3 // 78465 /// NM_ 1700084C01Rik BC150846 0.107391 -1.5916 0.0385418 -2.05801 0.295457 -1.29305 10569654 AK007416 // 1810010D01Rik // RIKEN cDNA 1810010D01 gene // 7 F5 // 381935 /// XR 1810010D01Rik AK007416 0.145576 1.69432 0.0476957 2.51662 0.288571 1.48533 10508883 NM_001083916 // 1810019J16Rik // RIKEN cDNA 1810019J16 gene // 4 D2.3 // 69073 / 1810019J16Rik NM_001083916 0.0533206 1.57139 0.0145433 2.56417 0.0836674 1.63179 10585282 ENSMUST00000050829 // 2010007H06Rik // RIKEN cDNA 2010007H06 gene // --- // 6984 2010007H06Rik ENSMUST00000050829 0.129914 -1.71998 0.0434862 -2.51672 -

Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 T + is online at: average * The Journal of Immunology , 34 of which you can access for free at: 2016; 197:1477-1488; Prepublished online 1 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600589 http://www.jimmunol.org/content/197/4/1477 Molecular Profile of Tumor-Specific CD8 Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh, Sonia M. Leach, Brandon L. Moore, Tullia C. Bruno, Jonathan D. Buhrman and Jill E. Slansky J Immunol cites 95 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2016/07/01/jimmunol.160058 9.DCSupplemental This article http://www.jimmunol.org/content/197/4/1477.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. The Journal of Immunology Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. -

Supplementary Data
Supplemental Data A novel mouse model of X-linked nephrogenic diabetes insipidus: Phenotypic analysis and therapeutic implications Jian Hua Li, Chung-Lin Chou, Bo Li, Oksana Gavrilova, Christoph Eisner, Jürgen Schnermann, Stasia A. Anderson, Chu-Xia Deng, Mark A. Knepper, and Jürgen Wess Supplemental Methods Metabolic cage studies. Animals were maintained in mouse metabolic cages (Hatteras Instruments, Cary, NC) under controlled temperature and light conditions (12 hr light and dark cycles). Mice received a fixed daily ration of 6.5 g of gelled diet per 20 g of body weight per day. The gelled diet was composed of 4 g of Basal Diet 5755 (Test Diet, Richmond, IN), 2.5 ml of deionized water, and 65 mg agar. Preweighted drinking water was provided ad libitum during the course of the study. Mice were acclimated in the metabolic cages for 1-2 days. Urine was collected under mineral oil in preweighted collection vials for successive 24 hr periods. Analysis of GPCR expression in mouse IMCD cells via TaqMan real-time qRT-PCR. Total RNA prepared from mouse IMCD tubule suspensions was reverse transcribed as described under Experimental Procedures. Tissues from ten 10-week old C57BL/6 WT mice were collected and pooled for each individual experiment. cDNA derived from 640 ng of RNA was mixed with an equal volume of TaqMan gene expression 2 x master mix (Applied Biosystems, Foster City, CA). 100 μl-aliquots of this mixture (corresponding to 80 ng of RNA) were added to each of the 8 fill ports of a 384-well plate of a mouse GPCR array panel (Applied Biosystems). -

Monitoring Nociception by Analyzing Gene Expression Changes in the Central Nervous System of Mice
Zurich Open Repository and Archive University of Zurich Main Library Strickhofstrasse 39 CH-8057 Zurich www.zora.uzh.ch Year: 2010 Monitoring nociception by analyzing gene expression changes in the central nervous system of mice Asner, I N Posted at the Zurich Open Repository and Archive, University of Zurich ZORA URL: https://doi.org/10.5167/uzh-46678 Dissertation Originally published at: Asner, I N. Monitoring nociception by analyzing gene expression changes in the central nervous system of mice. 2010, University of Zurich, Vetsuisse Faculty. Monitoring Nociception by Analyzing Gene Expression Changes in the Central Nervous System of Mice Dissertation zur Erlangung der naturwissenschaftlichen Doktorwürde (Dr. sc. nat) vorgelegt der Mathematisch-naturwissenschaftlichen Fakultät der Universität Zürich von Igor Asner von St. Cergue VD Promotionskomitee Prof. Dr. Peter Sonderegger Prof. Dr. Kurt Bürki Prof. Dr. Hanns Ulrich Zeilhofer Dr. Paolo Cinelli (Leitung der Dissertation) Zürich, 2010 Table of contents Table of content Curriculum vitae 6 Publications 9 Summary 11 Zusammenfassung 14 1. Introduction 17 1.1. Pain and nociception 17 1.1.1 Nociceptive neurons and Mechanoceptors 18 1.1.2 Activation of the nociceptive neurons at the periphery 21 1.1.2.1 Response to noxious heat 22 1.1.2.2 Response to noxious cold 23 1.1.2.3 Response to mechanical stress 24 1.1.3 Nociceptive message processing in the Spinal Cord 25 1.1.3.1 The lamina I and the ascending pathways 25 1.1.3.2 The lamina II and the descending pathways 26 1.1.4 Pain processing and integration in the brain 27 1.1.4.1 The Pain Matrix 27 1.1.4.2 Activation of the descending pathways 29 1.1.5 Inflammatory Pain 31 1.2. -

CD Markers Are Routinely Used for the Immunophenotyping of Cells
ptglab.com 1 CD MARKER ANTIBODIES www.ptglab.com Introduction The cluster of differentiation (abbreviated as CD) is a protocol used for the identification and investigation of cell surface molecules. So-called CD markers are routinely used for the immunophenotyping of cells. Despite this use, they are not limited to roles in the immune system and perform a variety of roles in cell differentiation, adhesion, migration, blood clotting, gamete fertilization, amino acid transport and apoptosis, among many others. As such, Proteintech’s mini catalog featuring its antibodies targeting CD markers is applicable to a wide range of research disciplines. PRODUCT FOCUS PECAM1 Platelet endothelial cell adhesion of blood vessels – making up a large portion molecule-1 (PECAM1), also known as cluster of its intracellular junctions. PECAM-1 is also CD Number of differentiation 31 (CD31), is a member of present on the surface of hematopoietic the immunoglobulin gene superfamily of cell cells and immune cells including platelets, CD31 adhesion molecules. It is highly expressed monocytes, neutrophils, natural killer cells, on the surface of the endothelium – the thin megakaryocytes and some types of T-cell. Catalog Number layer of endothelial cells lining the interior 11256-1-AP Type Rabbit Polyclonal Applications ELISA, FC, IF, IHC, IP, WB 16 Publications Immunohistochemical of paraffin-embedded Figure 1: Immunofluorescence staining human hepatocirrhosis using PECAM1, CD31 of PECAM1 (11256-1-AP), Alexa 488 goat antibody (11265-1-AP) at a dilution of 1:50 anti-rabbit (green), and smooth muscle KD/KO Validated (40x objective). alpha-actin (red), courtesy of Nicola Smart. PECAM1: Customer Testimonial Nicola Smart, a cardiovascular researcher “As you can see [the immunostaining] is and a group leader at the University of extremely clean and specific [and] displays Oxford, has said of the PECAM1 antibody strong intercellular junction expression, (11265-1-AP) that it “worked beautifully as expected for a cell adhesion molecule.” on every occasion I’ve tried it.” Proteintech thanks Dr. -

ADGRE2-NTF Is Regulated by Site- Specific N-Glycosylation
www.nature.com/scientificreports OPEN Membrane-association of EMR2/ ADGRE2-NTF is regulated by site- specifc N-glycosylation Received: 19 December 2017 Yi-Shu Huang1,4, Nien-Yi Chiang1, Gin-Wen Chang1 & Hsi-Hsien Lin1,2,3 Accepted: 27 February 2018 The evolutionarily conserved adhesion G protein-coupled receptors (aGPCRs) play critical roles in Published: xx xx xxxx biological processes as diverse as brain development, cell polarity and innate immune functions. A defning feature of aGPCRs is the GPCR autoproteolysis inducing (GAIN) domain capable of self- catalytic cleavage, resulting in the generation of an extracellular N-terminal fragment (NTF) and a seven-transmembrane C-terminal fragment (CTF) involved in the cellular adhesion and signaling functions, respectively. Interestingly, two diferent NTF subtypes have previously been identifed, namely an NTF that couples non-covalently with the CTF and a membrane-associated NTF that tethers on cell surface independently. The two NTF subtypes are expected to regulate aGPCR signaling via distinct mechanisms however their molecular characteristics are largely unknown. Herein, the membrane-associated NTF of EMR2/ADGRE2 is investigated and found to be modifed by diferential N-glycosylation. The membrane association of EMR2-NTF occurs in post-ER compartments and site- specifc N-glycosylation in the GAIN domain is involved in modulating its membrane-association ability. Finally, a unique amphipathic α-helix in the GAIN domain is identifed as a putative membrane anchor of EMR2-NTF. These results provide novel insights into the complex interaction and activation mechanisms of aGPCRs. Characterized by a long extracellular domain (ECD) with cell-adhesion functions and a seven-transmembrane (7TM) domain with signaling functions, the adhesion G protein-coupled receptors (aGPCRs) have been impli- cated in diverse biological activities and human diseases1. -

(12) United States Patent (10) Patent No.: US 9,005,612 B2 Ledbetter Et Al
US009005612B2 (12) United States Patent (10) Patent No.: US 9,005,612 B2 Ledbetter et al. (45) Date of Patent: *Apr. 14, 2015 (54) BINDING DOMAIN-IMMUNOGLOBULIN 5,455,030 A 10/1995 Ladner et al. FUSION PROTEINS $599. A 3: BestInsley et al. al. (75) Inventors: Jeffrey A. Ledbetter, Seattle, WA (US); 5,530,101 A 6/1996 Queen et al. Martha S. Hayden-Ledbetter, Seattle, 5,580,756. A 12/1996 Linsley et al. WA (US) 5,595,7215,585,089 A 1431/1997 SAS,Kaminski et al.1 (73) Assignee: Emergent Product Development 5,597,707 A 1/1997 Marken et al. Seattle, LLC, Seattle, WA (US) 3.W - A 3.87 EaCee ea. (*) Notice: Subject to any disclaimer, the term of this 5,645,835 A 7/1997 Fell, Jr. et al. patent is extended or adjusted under 35 $22.9 A ck 8. 3. Earl 530,387.1 U.S.C. 154(b) by 0 days. 5,693,762.w A 12/1997 QueenOCC et Cal. al. ............ This patent is Subject to a terminal dis- 5,709,859 A 1/1998 Aruffo et al. claimer 5,714,147 A 2, 1998 Capon et al. 5,721, 108 A 2f1998 Robinson et al. (21) Appl. No.: 13/451,641 5,736,137 A 4, 1998 Anderson et al. 5,770,197 A 6/1998 Linsley et al. (22) Filed: Apr. 20, 2012 5,773.253 A 6/1998 Linsley et al. O O 5,776.456 A 7/1998 Anderson et al. (65) Prior Publication Data 5,795,572 A 8/1998 Diegel et al. -

Atpase, Na+/K+ Transporting, Alpha 3 Polypeptide Homologous to 3'UTR
HUGO ID Name Nalm-6 TOM-1 Reh Karpas-422 DoHH -2 SU-DHL-5 Namalwa DG-75 Ramos Raji BEL EHEB BONNA-12 L-428 DEL BCP-1 BC-3 BCBL-1 JSC-1 PEL-SY HBL-6 DS-1 RPMI-8226 NCI-H929 L-363 SK-MM-2 ATP1A3 ATPase, Na+/K+ transporting, alpha 3 polypeptide CD24 homologous to 3'UTR of human CD24 gene ABCC5 multidrug resistance-associated protein (MRP5) CD72 CD72 antigen TCL1A Tcell leukemia/lymphoma 1 ITGB2 Integrin, beta 2 (antigen CD18 (p95)) ? nuclear ribonucleoprotein particle (hnRNP) SGT1 suppressor of G2 allele of skp1 homolog DNMT 1 DNA (cytosine-5-)-methyltransferase 1 GALE UDP-Galactose 4 epimerase (GALE) HADHSC L-3-hydroxyacyl-CoA dehydrogenase LIG4 DNA ligase IV LIG1 Ligase I, DNA, ATP-dependent CEBPG CCAA T/enhancer binding protein (C/EBP), gamma DCK Deoxycytidine kinase TCEA1 TRANSCRIPTION ELONGATION FACTOR S-II TCN 1 TRANSCOBALAMIN I PRECURSOR POLA2 DNA polymerase alpha subunit CCNG2 cyclin G2 RNPC1 Finkel-Biskis-Reilly murine sarcoma virus; Human seb4D RNPC1 Finkel-Biskis-Reilly murine sarcoma virus; Human seb4D DGKD Diacylglycerol kinase delta KIAA0220 Polycystic kidney disease protein 1 KIAA0220 calcium-dependent group X phospholipase A2 KIAA0220 calcium-dependent group X phospholipase A2 ALDH5A1 NAD+-dependent succinate-semialdehyde dehydrogenase CCNG2 Polycystic kidney disease 1 (autosomal dominant) PDCD4 nuclear antigen H731-like protein SSH3BP1 eps8 binding protein e3B1 MAP4K2 B lymphocyte serine/threonine protein kinase (GC kinase) MAPRE2 novel T-cell activation protein ZNFN1A Ikaros/LyF-1 homolog (hIk-1) FLJ22624 clone 23799 KIAA0355 -

New Structural Perspectives in G Protein-Coupled Receptor-Mediated Src Family Kinase Activation
International Journal of Molecular Sciences Review New Structural Perspectives in G Protein-Coupled Receptor-Mediated Src Family Kinase Activation Sandra Berndt * and Ines Liebscher Rudolf Schönheimer Institute of Biochemistry, Molecular Biochemistry, Medical Faculty, University of Leipzig, 04103 Leipzig, Germany; [email protected] * Correspondence: [email protected]; Tel.: +49-341-9722175 Abstract: Src family kinases (SFKs) are key regulators of cell proliferation, differentiation, and survival. The expression of these non-receptor tyrosine kinases is strongly correlated with cancer development and tumor progression. Thus, this family of proteins serves as an attractive drug target. The activation of SFKs can occur via multiple signaling pathways, yet many of them are poorly understood. Here, we summarize the current knowledge on G protein-coupled receptor (GPCR)- mediated regulation of SFKs, which is of considerable interest because GPCRs are among the most widely used pharmaceutical targets. This type of activation can occur through a direct interaction between the two proteins or be allosterically regulated by arrestins and G proteins. We postulate that a rearrangement of binding motifs within the active conformation of arrestin-3 mediates Src regulation by comparison of available crystal structures. Therefore, we hypothesize a potentially different activation mechanism compared to arrestin-2. Furthermore, we discuss the probable direct regulation of SFK by GPCRs and investigate the intracellular domains of exemplary GPCRs with conserved polyproline binding motifs that might serve as scaffolding domains to allow such a direct interaction. Large intracellular domains in GPCRs are often understudied and, in general, not much Citation: Berndt, S.; Liebscher, I. is known of their contribution to different signaling pathways. -
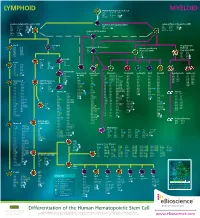
Lymphoid Myeloid
LYMPHOID Human Hematopoietic Stem Cell MYELOID CD34 CD117 (c-kit) CD338 CD38low/neg CD133 linneg CD59 CD135 (Flt3) GATA2 CD90 (Thy1) CD164 TdT Common Lymphoid Progenitor (CLP) Multi-Potent Progenitor (MPP) Common Myeloid Progenitor (CMP) low neg CD33 CD123low CD174 CD7 CD117 (c-kit) C/EBPα CD34 linneg neg CD34 CD131 Ikaros CD10 CD127 GATA2 CD135 (Flt3) TdT CD24neg CD135 (Flt3) GATA3 CD45RA CD164 PU.1 CD34 CD164 PU.1high Common DC Progenitor CD117 (c-kit) CD173 CD38 HLA-DR TdT CD45RA Aiolos CD11c dim CD90low c-mybhigh CD33 Pro-B Pre-NK/T Megakaryocyte- CD10 CD124 CD5 Conventional DC Precursor Erythroid CD19 CD164 CD34 CD11c Granulocyte-Myeloid CD20 CD252 CD14neg Progenitor (GMP) Progenitor (MEP) CD22 CD268 CD33dim CD33 CD131 CD34neg CD275 CD34 CD45RA CD34 CD164 CD72 HLA-DR CD36 FOG-1 CD74 CD123 PU.1 CD110 GATA1 CD123high GATA2 Pre-T Pro-NK CD7 Pre-B CD1a CD28 CD10neg CD2 CD34 neg CD10 CD124 CD34 CD3 CD45 CD117 (c-kit) CD19 CD164 CD5 CD127 (IL-7Rα) CD20 CD252 CD7high NOTCH1 CD22 CD268 CD34neg CD275 CD72 HLA-DR Immature NK CD74 IgM CD34neg Plasmacytoid Conventional Monocyte Neutrophil Eosinophil Mast Basophil Megakaryocyte Erythrocyte CD79a Pax5 CD94neg DC (pDC) DC (cDC) CD4low CD85a (ILT5) CD171 CD10 CD9 CD9 CD9 CD41 CD51 CD35 CD236 CD117 (c-kit) CD9 CD85d (ILT4) CD172a (SIRPα) CD15 CD11b CD11b CD11a CD42a CD61 CD44 CD236R CD4 CD1c CD122 CD11b CD85h (ILT1) CD172b CD16b CD15 CD15 CD11b CD42b CD110 CD123 CD238 CD11c neg CD1d CD161 CD11c CD85j (ILT2) CD180 CD17 CD24 CD24 CD13 CD42c CD112 CD173 CD239 CD45RA CD2 Ets-1 CD13 -

Human CD Marker Chart Reviewed by HLDA1 Bdbiosciences.Com/Cdmarkers
BD Biosciences Human CD Marker Chart Reviewed by HLDA1 bdbiosciences.com/cdmarkers 23-12399-01 CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD1a R4, T6, Leu6, HTA1 b-2-Microglobulin, CD74 + + + – + – – – CD93 C1QR1,C1qRP, MXRA4, C1qR(P), Dj737e23.1, GR11 – – – – – + + – – + – CD220 Insulin receptor (INSR), IR Insulin, IGF-2 + + + + + + + + + Insulin-like growth factor 1 receptor (IGF1R), IGF-1R, type I IGF receptor (IGF-IR), CD1b R1, T6m Leu6 b-2-Microglobulin + + + – + – – – CD94 KLRD1, Kp43 HLA class I, NKG2-A, p39 + – + – – – – – – CD221 Insulin-like growth factor 1 (IGF-I), IGF-II, Insulin JTK13 + + + + + + + + + CD1c M241, R7, T6, Leu6, BDCA1 b-2-Microglobulin + + + – + – – – CD178, FASLG, APO-1, FAS, TNFRSF6, CD95L, APT1LG1, APT1, FAS1, FASTM, CD95 CD178 (Fas ligand) + + + + + – – IGF-II, TGF-b latency-associated peptide (LAP), Proliferin, Prorenin, Plasminogen, ALPS1A, TNFSF6, FASL Cation-independent mannose-6-phosphate receptor (M6P-R, CIM6PR, CIMPR, CI- CD1d R3G1, R3 b-2-Microglobulin, MHC II CD222 Leukemia