2 #Run RENUM: Echo Renum.Par | Renumf90 | Tee Renum.Log # 3 #Cp Renf90.Par Pregs1.Par #Echo "OPTION Savecleansnps" &
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Software Developement Tools Introduction to Software Building
Software Developement Tools Introduction to software building SED & friends Outline 1. an example 2. what is software building? 3. tools 4. about CMake SED & friends – Introduction to software building 2 1 an example SED & friends – Introduction to software building 3 - non-portability: works only on a Unix systems, with mpicc shortcut and MPI libraries and headers installed in standard directories - every build, we compile all files - 0 level: hit the following line: mpicc -Wall -o heat par heat par.c heat.c mat utils.c -lm - 0.1 level: write a script (bash, csh, Zsh, ...) • drawbacks: Example • we want to build the parallel program solving heat equation: SED & friends – Introduction to software building 4 - non-portability: works only on a Unix systems, with mpicc shortcut and MPI libraries and headers installed in standard directories - every build, we compile all files - 0.1 level: write a script (bash, csh, Zsh, ...) • drawbacks: Example • we want to build the parallel program solving heat equation: - 0 level: hit the following line: mpicc -Wall -o heat par heat par.c heat.c mat utils.c -lm SED & friends – Introduction to software building 4 - non-portability: works only on a Unix systems, with mpicc shortcut and MPI libraries and headers installed in standard directories - every build, we compile all files • drawbacks: Example • we want to build the parallel program solving heat equation: - 0 level: hit the following line: mpicc -Wall -o heat par heat par.c heat.c mat utils.c -lm - 0.1 level: write a script (bash, csh, Zsh, ...) SED -

TEE Internal Core API Specification V1.1.2.50
GlobalPlatform Technology TEE Internal Core API Specification Version 1.1.2.50 (Target v1.2) Public Review June 2018 Document Reference: GPD_SPE_010 Copyright 2011-2018 GlobalPlatform, Inc. All Rights Reserved. Recipients of this document are invited to submit, with their comments, notification of any relevant patents or other intellectual property rights (collectively, “IPR”) of which they may be aware which might be necessarily infringed by the implementation of the specification or other work product set forth in this document, and to provide supporting documentation. The technology provided or described herein is subject to updates, revisions, and extensions by GlobalPlatform. This documentation is currently in draft form and is being reviewed and enhanced by the Committees and Working Groups of GlobalPlatform. Use of this information is governed by the GlobalPlatform license agreement and any use inconsistent with that agreement is strictly prohibited. TEE Internal Core API Specification – Public Review v1.1.2.50 (Target v1.2) THIS SPECIFICATION OR OTHER WORK PRODUCT IS BEING OFFERED WITHOUT ANY WARRANTY WHATSOEVER, AND IN PARTICULAR, ANY WARRANTY OF NON-INFRINGEMENT IS EXPRESSLY DISCLAIMED. ANY IMPLEMENTATION OF THIS SPECIFICATION OR OTHER WORK PRODUCT SHALL BE MADE ENTIRELY AT THE IMPLEMENTER’S OWN RISK, AND NEITHER THE COMPANY, NOR ANY OF ITS MEMBERS OR SUBMITTERS, SHALL HAVE ANY LIABILITY WHATSOEVER TO ANY IMPLEMENTER OR THIRD PARTY FOR ANY DAMAGES OF ANY NATURE WHATSOEVER DIRECTLY OR INDIRECTLY ARISING FROM THE IMPLEMENTATION OF THIS SPECIFICATION OR OTHER WORK PRODUCT. Copyright 2011-2018 GlobalPlatform, Inc. All Rights Reserved. The technology provided or described herein is subject to updates, revisions, and extensions by GlobalPlatform. -

Unix Tools 2
Unix Tools 2 Jeff Freymueller Outline § Variables as a collec9on of words § Making basic output and input § A sampler of unix tools: remote login, text processing, slicing and dicing § ssh (secure shell – use a remote computer!) § grep (“get regular expression”) § awk (a text processing language) § sed (“stream editor”) § tr (translator) § A smorgasbord of examples Variables as a collec9on of words § The shell treats variables as a collec9on of words § set files = ( file1 file2 file3 ) § This sets the variable files to have 3 “words” § If you want the whole variable, access it with $files § If you want just the second word, use $files[2] § The shell doesn’t count characters, only words Basic output: echo and cat § echo string § Writes a line to standard output containing the text string. This can be one or more words, and can include references to variables. § echo “Opening files” § echo “working on week $week” § echo –n “no carriage return at the end of this” § cat file § Sends the contents of a file to standard output Input, Output, Pipes § Output to file, vs. append to file. § > filename creates or overwrites the file filename § >> filename appends output to file filename § Take input from file, or from “inline input” § < filename take all input from file filename § <<STRING take input from the current file, using all lines un9l you get to the label STRING (see next slide for example) § Use a pipe § date | awk '{print $1}' Example of “inline input” gpsdisp << END • Many programs, especially Okmok2002-2010.disp Okmok2002-2010.vec older ones, interac9vely y prompt you to enter input Okmok2002-2010.gmtvec • You can automate (or self- y Okmok2002-2010.newdisp document) this by using << Okmok2002_mod.stacov • Standard input is set to the 5 contents of this file 5 Okmok2010.stacov between << END and END 5 • You can use any label, not 5 just “END”. -

Domain Tips and Tricks Lab
Installing a Domain Service for Windows: Domain Tips and Tricks Lab Novell Training Services www.novell.com OES10 ATT LIVE 2012 LAS VEGAS Novell, Inc. Copyright 2012-ATT LIVE-1-HARDCOPY PERMITTED. NO OTHER PRINTING, COPYING, OR DISTRIBUTION ALLOWED. Legal Notices Novell, Inc., makes no representations or warranties with respect to the contents or use of this documentation, and specifically disclaims any express or implied warranties of merchantability or fitness for any particular purpose. Further, Novell, Inc., reserves the right to revise this publication and to make changes to its content, at any time, without obligation to notify any person or entity of such revisions or changes. Further, Novell, Inc., makes no representations or warranties with respect to any software, and specifically disclaims any express or implied warranties of merchantability or fitness for any particular purpose. Further, Novell, Inc., reserves the right to make changes to any and all parts of Novell software, at any time, without any obligation to notify any person or entity of such changes. Any products or technical information provided under this Agreement may be subject to U.S. export controls and the trade laws of other countries. You agree to comply with all export control regulations and to obtain any required licenses or classification to export, re-export or import deliverables. You agree not to export or re-export to entities on the current U.S. export exclusion lists or to any embargoed or terrorist countries as specified in the U.S. export laws. You agree to not use deliverables for prohibited nuclear, missile, or chemical biological weaponry end uses. -

Show Command Output Redirection
show Command Output Redirection The show Command Output Redirection feature provides the capability to redirect output from Cisco IOS command-line interface (CLI) show commands and more commands to a file. • Finding Feature Information, page 1 • Information About show Command Output Redirection, page 1 • How to Use the show Command Enhancement, page 2 • Additional References, page 2 • Feature Information for show Command Output Redirection, page 3 Finding Feature Information Your software release may not support all the features documented in this module. For the latest caveats and feature information, see Bug Search Tool and the release notes for your platform and software release. To find information about the features documented in this module, and to see a list of the releases in which each feature is supported, see the feature information table at the end of this module. Use Cisco Feature Navigator to find information about platform support and Cisco software image support. To access Cisco Feature Navigator, go to www.cisco.com/go/cfn. An account on Cisco.com is not required. Information About show Command Output Redirection This feature enhances the show commands in the Cisco IOS CLI to allow large amounts of data output to be written directly to a file for later reference. This file can be saved on local or remote storage devices such as Flash, a SAN Disk, or an external memory device. For each show command issued, a new file can be created, or the output can be appended to an existing file. Command output can optionally be displayed on-screen while being redirected to a file by using the tee keyword. -

Useful Commands in Linux and Other Tools for Quality Control
Useful commands in Linux and other tools for quality control Ignacio Aguilar INIA Uruguay 05-2018 Unix Basic Commands pwd show working directory ls list files in working directory ll as before but with more information mkdir d make a directory d cd d change to directory d Copy and moving commands To copy file cp /home/user/is . To copy file directory cp –r /home/folder . to move file aa into bb in folder test mv aa ./test/bb To delete rm yy delete the file yy rm –r xx delete the folder xx Redirections & pipe Redirection useful to read/write from file !! aa < bb program aa reads from file bb blupf90 < in aa > bb program aa write in file bb blupf90 < in > log Redirections & pipe “|” similar to redirection but instead to write to a file, passes content as input to other command tee copy standard input to standard output and save in a file echo copy stream to standard output Example: program blupf90 reads name of parameter file and writes output in terminal and in file log echo par.b90 | blupf90 | tee blup.log Other popular commands head file print first 10 lines list file page-by-page tail file print last 10 lines less file list file line-by-line or page-by-page wc –l file count lines grep text file find lines that contains text cat file1 fiel2 concatenate files sort sort file cut cuts specific columns join join lines of two files on specific columns paste paste lines of two file expand replace TAB with spaces uniq retain unique lines on a sorted file head / tail $ head pedigree.txt 1 0 0 2 0 0 3 0 0 4 0 0 5 0 0 6 0 0 7 0 0 8 0 0 9 0 0 10 -

Basic Plotting Codes in R
The Basics of R for Windows We will use the data set timetrial.repeated.dat to learn some basic code in R for Windows. Commands will be shown in a different font, e.g., read.table, after the command line prompt, shown here as >. After you open R, type your commands after the prompt in what is called the Console window. Do not type the prompt, just the command. R stores the variables you create in a working directory. From the File menu, you need to first change the working directory to the directory where your data are stored. 1. Reading in data files Download the data file from www.biostat.umn.edu/~lynn/correlated.html to your local disk directory, go to File -> Change Directory to select that directory, and then read the data in: > contact = read.table(file="timetrial.repeated.dat", header=T) Note: if the first row of the data file had data instead of variable names, you would need to use header=F in the read.table command. Now you have named the data set contact, which you can then see in R just by typing its name: > contact or you can look at, for example, just the first 10 rows of the data set: > contact[1:10,] time sex age shape trial subj 1 2.68 M 31 Box 1 1 2 4.14 M 31 Box 2 1 3 7.22 M 31 Box 3 1 4 8.00 M 31 Box 4 1 5 7.09 M 30 Box 1 2 6 8.55 M 30 Box 2 2 7 8.79 M 30 Box 3 2 8 9.68 M 30 Box 4 2 9 6.05 M 30 Box 1 3 10 6.25 M 30 Box 2 3 This data set has 6 variables, one each in a column, where sex and shape are character valued. -
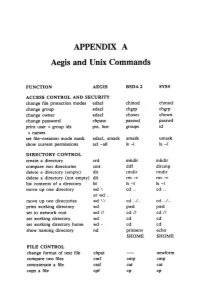
APPENDIX a Aegis and Unix Commands
APPENDIX A Aegis and Unix Commands FUNCTION AEGIS BSD4.2 SYSS ACCESS CONTROL AND SECURITY change file protection modes edacl chmod chmod change group edacl chgrp chgrp change owner edacl chown chown change password chpass passwd passwd print user + group ids pst, lusr groups id +names set file-creation mode mask edacl, umask umask umask show current permissions acl -all Is -I Is -I DIRECTORY CONTROL create a directory crd mkdir mkdir compare two directories cmt diff dircmp delete a directory (empty) dlt rmdir rmdir delete a directory (not empty) dlt rm -r rm -r list contents of a directory ld Is -I Is -I move up one directory wd \ cd .. cd .. or wd .. move up two directories wd \\ cd . ./ .. cd . ./ .. print working directory wd pwd pwd set to network root wd II cd II cd II set working directory wd cd cd set working directory home wd- cd cd show naming directory nd printenv echo $HOME $HOME FILE CONTROL change format of text file chpat newform compare two files emf cmp cmp concatenate a file catf cat cat copy a file cpf cp cp Using and Administering an Apollo Network 265 copy std input to std output tee tee tee + files create a (symbolic) link crl In -s In -s delete a file dlf rm rm maintain an archive a ref ar ar move a file mvf mv mv dump a file dmpf od od print checksum and block- salvol -a sum sum -count of file rename a file chn mv mv search a file for a pattern fpat grep grep search or reject lines cmsrf comm comm common to 2 sorted files translate characters tic tr tr SHELL SCRIPT TOOLS condition evaluation tools existf test test -

PS TEXT EDIT Reference Manual Is Designed to Give You a Complete Is About Overview of TEDIT
Information Management Technology Library PS TEXT EDIT™ Reference Manual Abstract This manual describes PS TEXT EDIT, a multi-screen block mode text editor. It provides a complete overview of the product and instructions for using each command. Part Number 058059 Tandem Computers Incorporated Document History Edition Part Number Product Version OS Version Date First Edition 82550 A00 TEDIT B20 GUARDIAN 90 B20 October 1985 (Preliminary) Second Edition 82550 B00 TEDIT B30 GUARDIAN 90 B30 April 1986 Update 1 82242 TEDIT C00 GUARDIAN 90 C00 November 1987 Third Edition 058059 TEDIT C00 GUARDIAN 90 C00 July 1991 Note The second edition of this manual was reformatted in July 1991; no changes were made to the manual’s content at that time. New editions incorporate any updates issued since the previous edition. Copyright All rights reserved. No part of this document may be reproduced in any form, including photocopying or translation to another language, without the prior written consent of Tandem Computers Incorporated. Copyright 1991 Tandem Computers Incorporated. Contents What This Book Is About xvii Who Should Use This Book xvii How to Use This Book xvii Where to Go for More Information xix What’s New in This Update xx Section 1 Introduction to TEDIT What Is PS TEXT EDIT? 1-1 TEDIT Features 1-1 TEDIT Commands 1-2 Using TEDIT Commands 1-3 Terminals and TEDIT 1-3 Starting TEDIT 1-4 Section 2 TEDIT Topics Overview 2-1 Understanding Syntax 2-2 Note About the Examples in This Book 2-3 BALANCED-EXPRESSION 2-5 CHARACTER 2-9 058059 Tandem Computers -

Installation Instruction
Rev 1/2020 Installation Instruction Please read the instructions completely before beginning the installation. Circ Collection Model Number: 3920CI Flush both supply lines before installation, then shut off both supply lines. Minimum hole size for the spout and handle trim is 1-1/4” and maximum of 1-1/2” SPOUT INSTALLATION 1 - Remove the tee connector, spout mounting hardware and tee connector washer from the bottom of the spout assembly. Apply a ring of silicon caulking around the center hole on the top of the sink. Do not use plumbers' putty which can damage the finish of the faucet. - From the top of the deck, insert the spout's threaded shank through the center hole of the sink. Make sure the spout flange washer is properly placed underneath of the flange and gently pressed against the sink top. Rubber washer 2 - Slip the spout mounting hardware back onto the spout. First the notched rubber washer, then the notched metal washer and lastly the brass lock nut. Hand tighten the lock nut. - Tighten the lock nut. Make sure the spout does not move side ways Adjust it if necessary. - Place the tee connector washer into the tee connector and thread the tee connector onto the end of the spout shank. Tighten to seal. - Align the spout and tighten the shank lock nut firmly. Notched rubber washer Notched metal washer Spout stabilizer Brass lock nut Tee connector washer Tee connector The measurements shown are for reference only. Products and specifications shown are subject to change without notice. www.santecfaucet.com | P: 310.542.0063 F: 310.542.5681 1/7 HANDLE TRIM INSTALLATION DO NOT DISASSEMBLE HANDLE ASSEMBLY - DROP-IN VALVES 1 - Both hot and cold side handle trims are pre-assmbled for faster installation. -

Keep TEE Probe Care Top-Of-Mind
Keep TEE Probe Care Top-Of-Mind Three Ways to Prevent Catastrophic TEE Probe Failures 1. Perform Frequent Quality Visual Inspections • When removed from storage • During removal • Before soaking in disinfectant • When set-up and connected to the scanner • After removal • After soaking in disinfectant • Before insertion • During pre-cleaning • Before storing 2. Perform Frequent and Time-based Leakage Testing Frequent • After EVERY patient exam, following a thorough visual inspection, under magnification, and prior to lengthy soak in disinfectant Time-based • Test once at the beginning of the soak cycle and once again at the end in order to reveal intermittent, slow leaks (the beginning of hard, catastrophic failures) 3. Establish a TEE Probe Preventative Maintenance Program • Address wearable items before they can contribute to larger, costly failures • Restore optimal performance and maintain uptime • Extend TEE probe lifecycyle • Suggested Intervals: • Bending rubber replacement - every 6-12 months • Articulation adjustment - every 6-12 months • Re-coating/re-labeling - every 12-18 months Industry Best-Practices for TEE Probe Care and Handling • Always use a protective tip cover when the probe is outside of the patient • Always transport TEE probes in covered bins • Don't coil the insertion tube in less than a 12-inch diameter • Use a TEE bite guard on every patient • Anytime a probe changes hands, perform a quick visual inspection of the tip, bending section and insertion tube. See below • Thoroughly inspect the tip and bending section, -

Attitudes Toward Disability
School of Psychological Science Attitudes Toward Disability: The Relationship Between Attitudes Toward Disability and Frequency of Interaction with People with Disabilities (PWD) # Jennifer Kim, Taylor Locke, Emily Reed, Jackie Yates, Nicholas Zike, Mariah Es?ll, BridgeBe Graham, Erika Frandrup, Kathleen Bogart, PhD, Sam Logan, PhD, Chris?na Hospodar Introduc)on Methods Con)nued Conclusion The social and medical models of disability are sets of underlying assump?ons explaining Parcipants Connued Suppor?ng our hypothesis, the medical and social models par?ally mediated the people's beliefs about the causes and implicaons of disability. • 7.9% iden?fied as Hispanic/Lano & 91.5% did not iden?fy as Hispanic/Lano relaonship between contact and atudes towards PWD. • 2.6% iden?fied as Black/African American, 1.7% iden?fied as American Indian or Alaska • Compared to the social model, the medical model was more strongly associated • The medical model is the predominant model in the United States that is associated Nave, 23.3% iden?fied as Asian, 2.2% iden?fied as Nave Hawaiian or Pacific Islander, with contact and atudes. with the belief that disability is an undesirable status that needs to be cured (Darling & 76.7% iden?fied as White, & 2.1% iden?fied as Other • This suggests that more contact with PWD can help decrease medical model beliefs Heckert, 2010). This model focuses on the diagnosis, treatment and curave efforts • Average frequency of contact was about once a month in general, but may be slightly less effec?ve at promo?ng the social model beliefs. related to disability.