C2ay26298c.Pdf
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genetic Landscape of Papillary Thyroid Carcinoma and Nuclear Architecture: an Overview Comparing Pediatric and Adult Populations
cancers Review Genetic Landscape of Papillary Thyroid Carcinoma and Nuclear Architecture: An Overview Comparing Pediatric and Adult Populations 1, 2, 2 3 Aline Rangel-Pozzo y, Luiza Sisdelli y, Maria Isabel V. Cordioli , Fernanda Vaisman , Paola Caria 4,*, Sabine Mai 1,* and Janete M. Cerutti 2 1 Cell Biology, Research Institute of Oncology and Hematology, University of Manitoba, CancerCare Manitoba, Winnipeg, MB R3E 0V9, Canada; [email protected] 2 Genetic Bases of Thyroid Tumors Laboratory, Division of Genetics, Department of Morphology and Genetics, Universidade Federal de São Paulo/EPM, São Paulo, SP 04039-032, Brazil; [email protected] (L.S.); [email protected] (M.I.V.C.); [email protected] (J.M.C.) 3 Instituto Nacional do Câncer, Rio de Janeiro, RJ 22451-000, Brazil; [email protected] 4 Department of Biomedical Sciences, University of Cagliari, 09042 Cagliari, Italy * Correspondence: [email protected] (P.C.); [email protected] (S.M.); Tel.: +1-204-787-2135 (S.M.) These authors contributed equally to this paper. y Received: 29 September 2020; Accepted: 26 October 2020; Published: 27 October 2020 Simple Summary: Papillary thyroid carcinoma (PTC) represents 80–90% of all differentiated thyroid carcinomas. PTC has a high rate of gene fusions and mutations, which can influence clinical and biological behavior in both children and adults. In this review, we focus on the comparison between pediatric and adult PTC, highlighting genetic alterations, telomere-related genomic instability and changes in nuclear organization as novel biomarkers for thyroid cancers. Abstract: Thyroid cancer is a rare malignancy in the pediatric population that is highly associated with disease aggressiveness and advanced disease stages when compared to adult population. -

Identification of Kinase Fusion Oncogenes in Post-Chernobyl Radiation-Induced Thyroid Cancers
Identification of kinase fusion oncogenes in post-Chernobyl radiation-induced thyroid cancers Julio C. Ricarte-Filho, … , Christopher E. Mason, James A. Fagin J Clin Invest. 2013;123(11):4935-4944. https://doi.org/10.1172/JCI69766. Research Article Exposure to ionizing radiation during childhood markedly increases the risk of developing papillary thyroid cancer. We examined tissues from 26 Ukrainian patients with thyroid cancer who were younger than 10 years of age and living in contaminated areas during the time of the Chernobyl nuclear reactor accident. We identified nonoverlapping somatic driver mutations in all 26 cases through candidate gene assays and next-generation RNA sequencing. We found that 22 tumors harbored fusion oncogenes that arose primarily through intrachromosomal rearrangements. Altogether, 23 of the oncogenic drivers identified in this cohort aberrantly activate MAPK signaling, including the 2 somatic rearrangements resulting in fusion of transcription factor ETS variant 6 (ETV6) with neurotrophic tyrosine kinase receptor, type 3 (NTRK3) and fusion of acylglycerol kinase (AGK) with BRAF. Two other tumors harbored distinct fusions leading to overexpression of the nuclear receptor PPARγ. Fusion oncogenes were less prevalent in tumors from a cohort of children with pediatric thyroid cancers that had not been exposed to radiation but were from the same geographical regions. Radiation-induced thyroid cancers provide a paradigm of tumorigenesis driven by fusion oncogenes that activate MAPK signaling or, less frequently, a PPARγ-driven transcriptional program. Find the latest version: https://jci.me/69766/pdf Related Commentary, page 4566 Research article Identification of kinase fusion oncogenes in post-Chernobyl radiation-induced thyroid cancers Julio C. -

1,2-Dimethylhydrazine-Induced Alterations in Protein Kinase C Activity in the Rat Preneoplastic Colon1
[CANCER RESEARCH 50. 3915-3920. July I. 1990) 1,2-Dimethylhydrazine-induced Alterations in Protein Kinase C Activity in the Rat Preneoplastic Colon1 Charles L. Baum, Ramesh K. Wali, Michael D. Sitrin, Merry J. G. Bolt, and Thomas A. Brasitus2 Department of Medicine, University of Chicago Hospitals and Clinics, Prit:ker School of Medicine, University of Chicago, Chicago, Illinois 60637 ABSTRACT which indicate that alterations in the activity of this kinase may also be involved in malignant transformation (7-14). Thus, Recently, a number of studies in experimental animals and humans tumor-promoting phorbol esters have been shown to bind to have suggested that alterations in the activity of protein kinase C (PKC) and directly activate PKC (7). Data indicate that PKC may also may be involved in the malignant transformation process. To determine play a role in the in vitro transformation processes produced by whether such alterations in this kinase were present before the develop a number of oncogenes (8-10). Cells that overproduce PKC ment of 1,2-dimethylhydrazine (DMH)-induced colon cancers, rats were given s.c. injections of this procarcinogen (20 mg/kg body weight/week) have, moreover, been shown to be more susceptible to transfor or diluent for 10 or 15 weeks. Animals were sacrificed after these time mation by one such activated oncogene, H-ras (11). Addition periods and colonie epithelium was harvested from each group. The ally, fibroblasts transfected with plasmids containing PKC- activity and distribution of PKC in the cytosolic and membrane fractions complementary DNA, overproduce PKC and demonstrate en of these preparations as well as 1,2-diacylglycerol mass and phosphoi- hanced tumorigenicity (12, 13). -

Downloaded on 02/15/2018)
proteomes Article Proteomic Analysis of 3T3-L1 Adipocytes Treated with Insulin and TNF-α 1, 2 1, 3 Hayley Chan y, Ketaki P. Bhide , Aditya Vaidyam z, Victoria Hedrick , Tiago Jose Paschoal Sobreira 3, Thomas G. Sors 4, Ryan W. Grant 5,§ and Uma K. Aryal 3,6,* 1 Department of Computer Science, Purdue University, West Lafayette, IN 47907, USA; [email protected] (H.C.); [email protected] (A.V.) 2 College of Agriculture, Purdue University, West Lafayette, IN 47907, USA; [email protected] 3 Purdue Proteomics Facility, Bindley Bioscience Center, Purdue University, West Lafayette, IN 47907, USA; [email protected] (V.H.); [email protected] (T.J.P.S.) 4 Purdue Institute of Inflammation, Immunology and Infectious Disease, Purdue University, West Lafayette, IN 47907, USA; [email protected] 5 Department of Nutrition Science, Purdue University, West Lafayette, IN 47907, USA; [email protected] 6 Department of Comparative Pathobiology, College of Veterinary Medicine, Purdue University, West Lafayette, IN 47907, USA * Correspondence: [email protected]; Tel.: +(765)-494-4960 Present addresses: Indiana University School of Medicine, West Lafayette, IN 47907, USA. y Present addresses: Division of Digital Psychiatry, Harvard Medical School, Boston, MA 02111, USA. z § Present addresses: Pharmavite, Los Angeles, CA 90089, USA. Received: 29 August 2019; Accepted: 17 October 2019; Published: 20 October 2019 Abstract: Insulin resistance is an indication of early stage Type 2 diabetes (T2D). Insulin resistant adipose tissues contain higher levels of insulin than the physiological level, as well as higher amounts of intracellular tumor necrosis factor-α (TNF-α) and other cytokines. However, the mechanism of insulin resistance remains poorly understood. -

Therapy Prospects for Mitochondrial DNA Maintenance Disorders
International Journal of Molecular Sciences Review Therapy Prospects for Mitochondrial DNA Maintenance Disorders Javier Ramón 1,2 , Ferran Vila-Julià 1,2, David Molina-Granada 1,2 , Miguel Molina-Berenguer 1,2 , Maria Jesús Melià 1,2 , Elena García-Arumí 1,2 , Javier Torres-Torronteras 1,2 , Yolanda Cámara 1,2 and Ramon Martí 1,2,* 1 Research Group on Neuromuscular and Mitochondrial Diseases, Vall d’Hebron Research Institute, Universitat Autònoma de Barcelona, 08035 Barcelona, Spain; [email protected] (J.R.); [email protected] (F.V.-J.); [email protected] (D.M.-G.); [email protected] (M.M.-B.); [email protected] (M.J.M.); [email protected] (E.G.-A.); [email protected] (J.T.-T.); [email protected] (Y.C.) 2 Biomedical Network Research Centre on Rare Diseases (CIBERER), Instituto de Salud Carlos III, 28029 Madrid, Spain * Correspondence: [email protected] Abstract: Mitochondrial DNA depletion and multiple deletions syndromes (MDDS) constitute a group of mitochondrial diseases defined by dysfunctional mitochondrial DNA (mtDNA) replication and maintenance. As is the case for many other mitochondrial diseases, the options for the treatment of these disorders are rather limited today. Some aggressive treatments such as liver transplantation or allogeneic stem cell transplantation are among the few available options for patients with some forms of MDDS. However, in recent years, significant advances in our knowledge of the biochemical pathomechanisms accounting for dysfunctional mtDNA replication have been achieved, which has opened new prospects for the treatment of these often fatal diseases. Current strategies under Citation: Ramón, J.; Vila-Julià, F.; investigation to treat MDDS range from small molecule substrate enhancement approaches to more Molina-Granada, D.; Molina-Berenguer, M.; Melià, M.J.; complex treatments, such as lentiviral or adenoassociated vector-mediated gene therapy. -

12) United States Patent (10
US007635572B2 (12) UnitedO States Patent (10) Patent No.: US 7,635,572 B2 Zhou et al. (45) Date of Patent: Dec. 22, 2009 (54) METHODS FOR CONDUCTING ASSAYS FOR 5,506,121 A 4/1996 Skerra et al. ENZYME ACTIVITY ON PROTEIN 5,510,270 A 4/1996 Fodor et al. MICROARRAYS 5,512,492 A 4/1996 Herron et al. 5,516,635 A 5/1996 Ekins et al. (75) Inventors: Fang X. Zhou, New Haven, CT (US); 5,532,128 A 7/1996 Eggers Barry Schweitzer, Cheshire, CT (US) 5,538,897 A 7/1996 Yates, III et al. s s 5,541,070 A 7/1996 Kauvar (73) Assignee: Life Technologies Corporation, .. S.E. al Carlsbad, CA (US) 5,585,069 A 12/1996 Zanzucchi et al. 5,585,639 A 12/1996 Dorsel et al. (*) Notice: Subject to any disclaimer, the term of this 5,593,838 A 1/1997 Zanzucchi et al. patent is extended or adjusted under 35 5,605,662 A 2f1997 Heller et al. U.S.C. 154(b) by 0 days. 5,620,850 A 4/1997 Bamdad et al. 5,624,711 A 4/1997 Sundberg et al. (21) Appl. No.: 10/865,431 5,627,369 A 5/1997 Vestal et al. 5,629,213 A 5/1997 Kornguth et al. (22) Filed: Jun. 9, 2004 (Continued) (65) Prior Publication Data FOREIGN PATENT DOCUMENTS US 2005/O118665 A1 Jun. 2, 2005 EP 596421 10, 1993 EP 0619321 12/1994 (51) Int. Cl. EP O664452 7, 1995 CI2O 1/50 (2006.01) EP O818467 1, 1998 (52) U.S. -

Dema and Faust Et Al., Suppl. Material 2020.02.03
Supplementary Materials Cyclin-dependent kinase 18 controls trafficking of aquaporin-2 and its abundance through ubiquitin ligase STUB1, which functions as an AKAP Dema Alessandro1,2¶, Dörte Faust1¶, Katina Lazarow3, Marc Wippich3, Martin Neuenschwander3, Kerstin Zühlke1, Andrea Geelhaar1, Tamara Pallien1, Eileen Hallscheidt1, Jenny Eichhorst3, Burkhard Wiesner3, Hana Černecká1, Oliver Popp1, Philipp Mertins1, Gunnar Dittmar1, Jens Peter von Kries3, Enno Klussmann1,4* ¶These authors contributed equally to this work 1Max Delbrück Center for Molecular Medicine in the Helmholtz Association (MDC), Robert- Rössle-Strasse 10, 13125 Berlin, Germany 2current address: University of California, San Francisco, 513 Parnassus Avenue, CA 94122 USA 3Leibniz-Forschungsinstitut für Molekulare Pharmakologie (FMP), Robert-Rössle-Strasse 10, 13125 Berlin, Germany 4DZHK (German Centre for Cardiovascular Research), Partner Site Berlin, Oudenarder Strasse 16, 13347 Berlin, Germany *Corresponding author Enno Klussmann Max Delbrück Center for Molecular Medicine Berlin in the Helmholtz Association (MDC) Robert-Rössle-Str. 10, 13125 Berlin Germany Tel. +49-30-9406 2596 FAX +49-30-9406 2593 E-mail: [email protected] 1 Content 1. CELL-BASED SCREENING BY AUTOMATED IMMUNOFLUORESCENCE MICROSCOPY 3 1.1 Screening plates 3 1.2 Image analysis using CellProfiler 17 1.4 Identification of siRNA affecting cell viability 18 1.7 Hits 18 2. SUPPLEMENTARY TABLE S4, FIGURES S2-S4 20 2 1. Cell-based screening by automated immunofluorescence microscopy 1.1 Screening plates Table S1. Genes targeted with the Mouse Protein Kinases siRNA sub-library. Genes are sorted by plate and well. Accessions refer to National Center for Biotechnology Information (NCBI, BLA) entries. The siRNAs were arranged on three 384-well microtitre platres. -

Autocrine IFN Signaling Inducing Profibrotic Fibroblast Responses By
Downloaded from http://www.jimmunol.org/ by guest on September 23, 2021 Inducing is online at: average * The Journal of Immunology , 11 of which you can access for free at: 2013; 191:2956-2966; Prepublished online 16 from submission to initial decision 4 weeks from acceptance to publication August 2013; doi: 10.4049/jimmunol.1300376 http://www.jimmunol.org/content/191/6/2956 A Synthetic TLR3 Ligand Mitigates Profibrotic Fibroblast Responses by Autocrine IFN Signaling Feng Fang, Kohtaro Ooka, Xiaoyong Sun, Ruchi Shah, Swati Bhattacharyya, Jun Wei and John Varga J Immunol cites 49 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2013/08/20/jimmunol.130037 6.DC1 This article http://www.jimmunol.org/content/191/6/2956.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 23, 2021. The Journal of Immunology A Synthetic TLR3 Ligand Mitigates Profibrotic Fibroblast Responses by Inducing Autocrine IFN Signaling Feng Fang,* Kohtaro Ooka,* Xiaoyong Sun,† Ruchi Shah,* Swati Bhattacharyya,* Jun Wei,* and John Varga* Activation of TLR3 by exogenous microbial ligands or endogenous injury-associated ligands leads to production of type I IFN. -
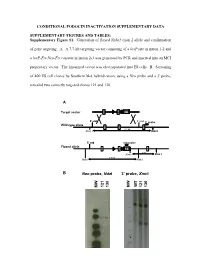
Supplementary Figure S1. Generation of Floxed Nphs2 Exon 2 Allele and Confirmation
CONDITIONAL PODOCIN INACTIVATION SUPPLEMENTARY DATA SUPPLEMENTARY FIGURES AND TABLES: Supplementary Figure S1. Generation of floxed Nphs2 exon 2 allele and confirmation of gene targeting. A. A 7.7-kb targeting vector consisting of a loxP site in intron 1-2 and a loxP-Frt-Neo-Frt cassette in intron 2-3 was generated by PCR and inserted into an MCI proprietary vector. The linearized vector was electroporated into ES cells. B. Screening of 400 ES cell clones by Southern blot hybridization, using a Neo probe and a 3' probe, revealed two correctly targeted clones 121 and 130. A Target vector 1 2 NEO 5’ end 3’ end 3’ probe Wild-type allele 1 2 3 9.2 kb Xmn I Xmn I 5’ end neo probe Floxed allele 1 2 NEO 3 5 kb Xmn I Xmn I 7.8 kb Nde I Nde I B Neo probe, NdeI 3’ probe, XmnI WT 121 130 121 130 MW MW - 1 - Supplementary Figure S2. Generation of Nphs2lox2/-,Cre+ mice and excision of exon 2 upon Cre recombinase induction. A. Triallelic Nphs2lox2/-,Cre+ mice were obtained by mating phenotypically normal Nphs2lox2/lox2 mice with Nphs2+/-,Cre+ mice. Mendelian inheritance of these alleles was observed. B. Genotypes were verified by multiplex PCR of tail genomic DNA. C. Cre recombinase activity was induced upon tamoxifen administration, leading to excision of the floxed exon 2 of the Nphs2 gene. D. Cre recombinase activity in the kidney was verified by PCR using a set of forward and reverse primers designed around exon 2 and demonstrating a 692-bp product (before Cre) and a 316-bp product (after Cre) using genomic DNA extracted from the renal cortex. -

Supplemental Figures 04 12 2017
Jung et al. 1 SUPPLEMENTAL FIGURES 2 3 Supplemental Figure 1. Clinical relevance of natural product methyltransferases (NPMTs) in brain disorders. (A) 4 Table summarizing characteristics of 11 NPMTs using data derived from the TCGA GBM and Rembrandt datasets for 5 relative expression levels and survival. In addition, published studies of the 11 NPMTs are summarized. (B) The 1 Jung et al. 6 expression levels of 10 NPMTs in glioblastoma versus non‐tumor brain are displayed in a heatmap, ranked by 7 significance and expression levels. *, p<0.05; **, p<0.01; ***, p<0.001. 8 2 Jung et al. 9 10 Supplemental Figure 2. Anatomical distribution of methyltransferase and metabolic signatures within 11 glioblastomas. The Ivy GAP dataset was downloaded and interrogated by histological structure for NNMT, NAMPT, 12 DNMT mRNA expression and selected gene expression signatures. The results are displayed on a heatmap. The 13 sample size of each histological region as indicated on the figure. 14 3 Jung et al. 15 16 Supplemental Figure 3. Altered expression of nicotinamide and nicotinate metabolism‐related enzymes in 17 glioblastoma. (A) Heatmap (fold change of expression) of whole 25 enzymes in the KEGG nicotinate and 18 nicotinamide metabolism gene set were analyzed in indicated glioblastoma expression datasets with Oncomine. 4 Jung et al. 19 Color bar intensity indicates percentile of fold change in glioblastoma relative to normal brain. (B) Nicotinamide and 20 nicotinate and methionine salvage pathways are displayed with the relative expression levels in glioblastoma 21 specimens in the TCGA GBM dataset indicated. 22 5 Jung et al. 23 24 Supplementary Figure 4. -

Impact of Exome Sequencing in Characterizing Mendelian Disorders
Journal of Human Genetics (2012) 57, 621–632 & 2012 The Japan Society of Human Genetics All rights reserved 1434-5161/12 $32.00 www.nature.com/jhg REVIEW Next-generation sequencing: impact of exome sequencing in characterizing Mendelian disorders Bahareh Rabbani1, Nejat Mahdieh1, Kazuyoshi Hosomichi, Hirofumi Nakaoka and Ituro Inoue Traditional approaches for gene mapping from candidate gene studies to positional cloning strategies have been applied for Mendelian disorders. Since 2005, next-generation sequencing (NGS) technologies are improving as rapid, high-throughput and cost-effective approaches to fulfill medical sciences and research demands. Using NGS, the underlying causative genes are directly distinguished via a systematic filtering, in which the identified gene variants are checked for novelty and functionality. During the past 2 years, the role of more than 100 genes has been distinguished in rare Mendelian disorders by means of whole-exome sequencing (WES). Combination of WES with traditional approaches, consistent with linkage analysis, has had the greatest impact on those disorders following autosomal mode of inheritance; in more than 60 identified genes, the causal variants have been transmitted at homozygous or compound heterozygous state. Recent literatures focusing on identified new causal genes in Mendelian disorders using WES are reviewed in the present survey. Journal of Human Genetics (2012) 57, 621–632; doi:10.1038/jhg.2012.91; published online 26 July 2012 Keywords: exome sequencing; mendelian disorder; mutation; next-generation sequencing; NGS; WES INTRODUCTION genes and 4535 disorders with known molecular basis (http:// One of the major concerns of medical sciences is finding the causal omim.org/statistics/geneMap). -

1787 Genome-Wide Identification and Characterization of Eukaryotic Protein Kinases Kanhu Charan Das1, Parismita Kalita1, Timir
[Frontiers in Bioscience, Landmark, 25, 1787-1827, June 1, 2020] Genome-wide identification and characterization of eukaryotic protein kinases Kanhu Charan Das1, Parismita Kalita1, Timir Tripathi1 1Molecular and Structural Biophysics Laboratory, Department of Biochemistry, North-Eastern Hill University, Shillong 793022, India TABLE OF CONTENTS 1. Abstract 2. Introduction 3. Methods 3.1. F. gigantica gene sequence retrieval 3.2. Generation of hidden markov models (HMMs) 3.3. Classification and domain analysis of FgePKs 4. Results 4.1. Identification of kinase-like proteins in F. gigantica genome 4.2. Classification and domain analysis of FgePKs 4.2.1. Ser/Thr kinase group 4.2.1.1. cAMP-dependent protein kinase (AGC) group 4.2.1.1.1. Protein kinase C (PKC) 4.2.1.1.2. Protein kinase A (PKA) 4.2.1.2. Calmodulin kinase (CaMK) group 4.2.1.3. Cyclin-dependent kinase and other close relatives (CMGC) group 4.2.1.3.1. Cyclin dependent kinases (CDK) 4.2.1.3.2. Mitogen activated protein kinases (MAPK) 4.2.1.3.3. Glycogen synthase 3 kinases (GSK3) 4.2.1.3.4. Dual-specificity Tyr regulated kinases (DYRK) 4.2.1.4. Sterile Ser/Thr kinase (STE) group 4.2.1.5. Casein kinase 1 (CK1) group 4.2.2. Tyr kinase group 4.3.3. Hybrid group 4.3.3.1. Tyr kinase-like (TKL) group 4.3.3.2. Others 5. Discussion 6. Acknowledgments 7. References 1. ABSTRACT The tropical liver fluke, Fasciola gigantica is kinases (ePKs) are regulators of cellular a food-borne parasite responsible for the phosphorylation.