Cell Biology
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Imaging, Tracking and Computational Analyses of Virus Entry and Egress
1 Review 2 Imaging, Tracking and Computational Analyses of 3 Virus Entry and Egress with the Cytoskeleton 4 I-Hsuan Wang 1,†, Christoph J. Burckhardt 2,†, A. Yakimovich 3 and Urs F. Greber 4,* 5 1 Division of Virology, Institute of Medical Science, tHe University of ToKyo, ToKyo 108-8639, Japan 6 2 UT SoutHwestern Medical Center, Lyda Hill Department of Bioinformatics, Dallas TX 75390, USA 7 3 MRC Laboratory for Molecular Cell Biology, University College London, London, United Kingdom 8 4 Department of Molecular Life Sciences, University of ZuricH, WintertHurerstrasse 190, CH-8057 ZuricH, 9 Switzerland 10 * Correspondence: [email protected], Telephone: +41 44 635 4841, Fax: +41 44 635 6817 11 † These autHors contributed equally to tHis work. 12 Received: date; Accepted: date; PublisHed: date 13 Abstract: Viruses Have a dual nature - particles are ‘passive substances’ lacKing chemical energy 14 transformation, wHereas infected cells are ‘active substances’ turning-over energy. How passive 15 viral substances convert to active substances, comprising viral replication and assembly 16 compartments Has been of intense interest to virologists, cell and molecular biologists and 17 immunologists. Infection starts witH virus entry into a susceptible cell and delivers tHe viral 18 genome to the replication site. THis is a multi-step process, and involves tHe cytosKeleton and 19 associated motor proteins. LiKewise, the egress of progeny virus particles from the replication site 20 to the extracellular space is enHanced by tHe cytosKeleton and associated motor proteins. THis 21 overcomes tHe limitation of tHermal diffusion, and transports virions and virion components, 22 often in association witH cellular organelles. -

P1 Bacteriophage and Tol System Mutants
P1 BACTERIOPHAGE AND TOL SYSTEM MUTANTS Cari L. Smerk A Thesis Submitted to the Graduate College of Bowling Green State University in partial fulfillment of the requirements for the degree of MASTER OF SCIENCE August 2007 Committee: Ray A. Larsen, Advisor Tami C. Steveson Paul A. Moore Lee A. Meserve ii ABSTRACT Dr. Ray A. Larsen, Advisor The integrity of the outer membrane of Gram negative bacteria is dependent upon proteins of the Tol system, which transduce cytoplasmic-membrane derived energy to as yet unidentified outer membrane targets (Vianney et al., 1996). Mutations affecting the Tol system of Escherichia coli render the cells resistant to a bacteriophage called P1 by blocking the phage maturation process in some way. This does not involve outer membrane interactions, as a mutant in the energy transucer (TolA) retained wild type levels of phage sensitivity. Conversely, mutations affecting the energy harvesting complex component, TolQ, were resistant to lysis by bacteriophage P1. Further characterization of specific Tol system mutants suggested that phage maturation was not coupled to energy transduction, nor to infection of the cells by the phage. Quantification of the number of phage produced by strains lacking this protein also suggests that the maturation of P1 phage requires conditions influenced by TolQ. This study aims to identify the role that the TolQ protein plays in the phage maturation process. Strains of cells were inoculated with bacteriophage P1 and the resulting production by the phage of viable progeny were determined using one step growth curves (Ellis and Delbruck, 1938). Strains that were lacking the TolQ protein rendered P1 unable to produce the characteristic burst of progeny phage after a single generation of phage. -

An Expanded View of Viruses
An Expanded View of Viruses Urs F. Greber 1) & Ralf Bartenschlager 2) 1) Department of Molecular Life Sciences, University of Zurich, Winterthurerstrasse 190, 8057 Zurich, Switzerland 2) Department of Infectious Diseases, Molecular Virology, Heidelberg University, Im Neuenheimer Feld 345, 69198 Heidelberg, Germany Correspondence to: [email protected] [email protected] 1 figure Keywords: Zoonosis; Pandemics; Dengue, Ebola; Influenza; Entry; Endocytosis; Uncoating; Transport; Filovirus; Comparative genomics; Computational analyses; Virus-host interactions; Filamentous virus; Bacterial biofilm; Phage; Bacteriophage; Glycan; Immunity; Infection; Disease; Evolution; Variability; Giant virus; Ferret; Guinea pig; Emerging disease; Pandemic; Host range; Enveloped virus; 1 Viruses are ubiquitous, and are important in medicine, biology, biotechnology and ecology. All kinds of cells can be infected with viruses, and sometimes, a particular cell is infected with different viruses at the same time. The virus particle, ‘virion’ is composed of the viral coat proteins sheltering the viral genome, and is often surrounded by a lipid “envelope”. A virion is small compared to cells, and when it enters cells gives rise to infection distinct from an intracellular bacterial pathogen (Lwoff, 1957). A virus-infected cell has a profoundly altered homeostasis due to numerous interactions between cellular and viral components. This leads to evolutionary pressure on both virus and host, and argues that viruses are a part of life (Ludmir & Enquist, 2009). Our cells can be infected by viruses causing acute disease, such as respiratory disease by Influenza virus or rhinoviruses, or chronic disease, such as hepatitis or immune deficiency. However, most viral attacks on cells are fend off, or the spread of viruses in an infected organism is restricted, and infection abrogated. -

Persistent Virus and Addiction Modules: an Engine of Symbiosis
UC Irvine UC Irvine Previously Published Works Title Persistent virus and addiction modules: an engine of symbiosis. Permalink https://escholarship.org/uc/item/5ck1g026 Journal Current opinion in microbiology, 31 ISSN 1369-5274 Author Villarreal, Luis P Publication Date 2016-06-01 DOI 10.1016/j.mib.2016.03.005 Peer reviewed eScholarship.org Powered by the California Digital Library University of California Available online at www.sciencedirect.com ScienceDirect Persistent virus and addiction modules: an engine of symbiosis Luis P Villarreal The giant DNA viruses are highly prevalent and have a particular host would occasionally survive but still retain a bit of affinity for the lytic infection of unicellular eukaryotic host. The the selfish virus DNA. Thus although parasitic selfish giant viruses can also be infected by inhibitory virophage which (virus-like) information is common in the genomes of all can provide lysis protection to their host. The combined life forms, its presence was explained as mostly defective protective and destructive action of such viruses can define a remnants of past plague sweeps that provides no func- general model (PD) of virus-mediated host survival. Here, I tional benefit to the host (e.g. junk). Until recently, this present a general model for role such viruses play in the explanation seemed satisfactory. In the last twenty years, evolution of host symbiosis. By considering how virus mixtures however, various observation-based developments have can participate in addiction modules, I provide a functional compelled us to re-evaluate this stance. Both comparative explanation for persistence of virus derived genetic ‘junk’ in genomics and metagenomics (sequencing habitats) has their host genomic habitats. -

Structural Variability and Complexity of the Giant Pithovirus Sibericum
www.nature.com/scientificreports OPEN Structural variability and complexity of the giant Pithovirus sibericum particle revealed by high- Received: 29 March 2017 Accepted: 22 September 2017 voltage electron cryo-tomography Published: xx xx xxxx and energy-fltered electron cryo- microscopy Kenta Okamoto1, Naoyuki Miyazaki2, Chihong Song2, Filipe R. N. C. Maia1, Hemanth K. N. Reddy1, Chantal Abergel 3, Jean-Michel Claverie3,4, Janos Hajdu1,5, Martin Svenda1 & Kazuyoshi Murata2 The Pithoviridae giant virus family exhibits the largest viral particle known so far, a prolate spheroid up to 2.5 μm in length and 0.9 μm in diameter. These particles show signifcant variations in size. Little is known about the structure of the intact virion due to technical limitations with conventional electron cryo-microscopy (cryo-EM) when imaging thick specimens. Here we present the intact structure of the giant Pithovirus sibericum particle at near native conditions using high-voltage electron cryo- tomography (cryo-ET) and energy-fltered cryo-EM. We detected a previously undescribed low-density outer layer covering the tegument and a periodical structuring of the fbres in the striated apical cork. Energy-fltered Zernike phase-contrast cryo-EM images show distinct substructures inside the particles, implicating an internal compartmentalisation. The density of the interior volume of Pithovirus particles is three quarters lower than that of the Mimivirus. However, it is remarkably high given that the 600 kbp Pithovirus genome is only half the size of the Mimivirus genome and is packaged in a volume up to 100 times larger. These observations suggest that the interior is densely packed with macromolecules in addition to the genomic nucleic acid. -

Structural Variability and Complexity of the Giant Pithovirus Sibericum
Structural variability and complexity of the giant Pithovirus sibericum particle revealed by high-voltage electron cryo-tomography and energy-filtered electron cryo-microscopy Kenta Okamoto, Naoyuki Miyazaki, Chihong Song, Filipe Maia, Hemanth Reddy, Chantal Abergel, Jean-Michel Claverie, Janos Hajdu, Martin Svenda, Kazuyoshi Murata To cite this version: Kenta Okamoto, Naoyuki Miyazaki, Chihong Song, Filipe Maia, Hemanth Reddy, et al.. Structural variability and complexity of the giant Pithovirus sibericum particle revealed by high-voltage electron cryo-tomography and energy-filtered electron cryo-microscopy. Scientific Reports, Nature Publishing Group, 2017, 7 (1), pp.13291. 10.1038/s41598-017-13390-4. hal-01785112 HAL Id: hal-01785112 https://hal-amu.archives-ouvertes.fr/hal-01785112 Submitted on 4 May 2018 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. www.nature.com/scientificreports OPEN Structural variability and complexity of the giant Pithovirus sibericum particle revealed by high- Received: 29 March 2017 Accepted: 22 September 2017 voltage electron cryo-tomography Published: xx xx xxxx and energy-fltered electron cryo- microscopy Kenta Okamoto1, Naoyuki Miyazaki2, Chihong Song2, Filipe R. N. C. Maia1, Hemanth K. N. Reddy1, Chantal Abergel 3, Jean-Michel Claverie3,4, Janos Hajdu1,5, Martin Svenda1 & Kazuyoshi Murata2 The Pithoviridae giant virus family exhibits the largest viral particle known so far, a prolate spheroid up to 2.5 μm in length and 0.9 μm in diameter. -

Herpesviral Latency—Common Themes
pathogens Review Herpesviral Latency—Common Themes Magdalena Weidner-Glunde * , Ewa Kruminis-Kaszkiel and Mamata Savanagouder Department of Reproductive Immunology and Pathology, Institute of Animal Reproduction and Food Research of Polish Academy of Sciences, Tuwima Str. 10, 10-748 Olsztyn, Poland; [email protected] (E.K.-K.); [email protected] (M.S.) * Correspondence: [email protected] Received: 22 January 2020; Accepted: 14 February 2020; Published: 15 February 2020 Abstract: Latency establishment is the hallmark feature of herpesviruses, a group of viruses, of which nine are known to infect humans. They have co-evolved alongside their hosts, and mastered manipulation of cellular pathways and tweaking various processes to their advantage. As a result, they are very well adapted to persistence. The members of the three subfamilies belonging to the family Herpesviridae differ with regard to cell tropism, target cells for the latent reservoir, and characteristics of the infection. The mechanisms governing the latent state also seem quite different. Our knowledge about latency is most complete for the gammaherpesviruses due to previously missing adequate latency models for the alpha and beta-herpesviruses. Nevertheless, with advances in cell biology and the availability of appropriate cell-culture and animal models, the common features of the latency in the different subfamilies began to emerge. Three criteria have been set forth to define latency and differentiate it from persistent or abortive infection: 1) persistence of the viral genome, 2) limited viral gene expression with no viral particle production, and 3) the ability to reactivate to a lytic cycle. This review discusses these criteria for each of the subfamilies and highlights the common strategies adopted by herpesviruses to establish latency. -

Bacterial Virus Ontology; Coordinating Across Databases
viruses Article Bacterial Virus Ontology; Coordinating across Databases Chantal Hulo 1, Patrick Masson 1, Ariane Toussaint 2, David Osumi-Sutherland 3, Edouard de Castro 1, Andrea H. Auchincloss 1, Sylvain Poux 1, Lydie Bougueleret 1, Ioannis Xenarios 1 and Philippe Le Mercier 1,* 1 Swiss-Prot group, SIB Swiss Institute of Bioinformatics, CMU, University of Geneva Medical School, 1211 Geneva, Switzerland; [email protected] (C.H.); [email protected] (P.M.); [email protected] (E.d.C.); [email protected] (A.H.A.); [email protected] (S.P.); [email protected] (L.B.); [email protected] (I.X.) 2 University Libre de Bruxelles, Génétique et Physiologie Bactérienne (LGPB), 12 rue des Professeurs Jeener et Brachet, 6041 Charleroi, Belgium; [email protected] 3 European Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI), Wellcome Trust Genome Campus, Hinxton CB10 1SD, UK; [email protected] * Correspondence: [email protected]; Tel.: +41-22379-5870 Academic Editors: Tessa E. F. Quax, Matthias G. Fischer and Laurent Debarbieux Received: 13 April 2017; Accepted: 17 May 2017; Published: 23 May 2017 Abstract: Bacterial viruses, also called bacteriophages, display a great genetic diversity and utilize unique processes for infecting and reproducing within a host cell. All these processes were investigated and indexed in the ViralZone knowledge base. To facilitate standardizing data, a simple ontology of viral life-cycle terms was developed to provide a common vocabulary for annotating data sets. New terminology was developed to address unique viral replication cycle processes, and existing terminology was modified and adapted. -
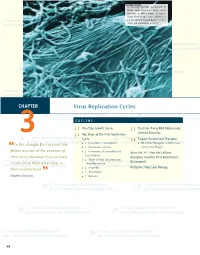
Virus Replication Cycles
© Jones and Bartlett Publishers. NOT FOR SALE OR DISTRIBUTION A scanning electron micrograph of Ebola virus particles. Ebola virus contains an RNA genome. It causes Ebola hemorrhagic fever, which is a severe and often fatal disease in hu- mans and nonhuman primates. CHAPTER Virus Replication Cycles OUTLINE 3.1 One-Step Growth Curves 3.3 The Error-Prone RNA Polymerases: 3 3.2 Key Steps of the Viral Replication Genetic Diversity Cycle 3.4 Targets for Antiviral Therapies In the struggle for survival, the ■ 1. Attachment (Adsorption) ■ RNA Virus Mutagens: A New Class “ ■ 2. Penetration (Entry) of Antiviral Drugs? fi ttest win out at the expense of ■ 3. Uncoating (Disassembly and Virus File 3-1: How Are Cellular Localization) their rivals because they succeed Receptors Used for Viral Attachment ■ 4. Types of Viral Genomes and Discovered? in adapting themselves best to Their Replication their environment. ■ 5. Assembly Refresher: Molecular Biology ” ■ 6. Maturation Charles Darwin ■ 7. Release 46 229329_CH03_046_069.indd9329_CH03_046_069.indd 4466 11/18/08/18/08 33:19:08:19:08 PPMM © Jones and Bartlett Publishers. NOT FOR SALE OR DISTRIBUTION CASE STUDY The campus day care was recently closed during the peak of the winter fl u season because many of the young children were sick with a lower respiratory tract infection. An email an- nouncement was sent to all students, faculty, and staff at the college that stated the closure was due to a metapneumovirus outbreak. The announcement briefed the campus com- munity with information about human metapneumonoviruses (hMPVs). The announcement stated that hMPV was a newly identifi ed respiratory tract pathogen discovered in the Netherlands in 2001. -

Viruses Chapter 19
Viruses Chapter 19 What you must know: The components of a virus. The differences between lytic and lysogenic cycles. How viruses can introduce genetic variation into host organisms. Mechanisms that introduce genetic variation into viral populations. Bacteria vs. Viruses Bacteria Virus Prokaryotic cell Not a living cell (genes Most are free-living (some packaged in protein shell) parasitic) Intracellular parasite Relatively large size 1/1000 size of bacteria Antibiotics used to kill Vaccines used to prevent bacteria viral infection Antiviral treatment Viruses Very small (<ribosomes) Components = nucleic acid + capsid ◦ Nucleic acid: DNA or RNA (double or single-stranded) ◦ Capsid: protein shell ◦ Some viruses also have viral envelopes that surround capsid Derived from host cell membranes and viral proteins Viruses Limited host range ◦ Entry = attach to host cell membrane receptors through capsid proteins or glycoproteins on viral envelope (animal) ◦ Eg. human cold virus (rhinovirus) upper respiratory tract (mouth & nose) Reproduce quickly within host cells Can mutate easily ◦ RNA viruses: no error-checking mechanisms Simplified viral replicative cycle Entry: Injection endocytosis, fusion VIDEO: T4 PHAGE INFECTION Bacteriophage Virus that infects bacterial cells Viral Reproduction - phages Lytic Cycle: ◦ Use host machinery to replicate, assemble, and release copies of virus ◦ Virulent phages: Cells die through lysis or apoptosis Lysogenic (Latent) Cycle: ◦ DNA incorporated into host DNA and replicated along with it -

30,000 Year-Old Giant Virus Found in Siberia
NATIONAL PRESS RELEASE I PARIS I MARCH 3, 2014 30,000 year-old giant virus found in Siberia A new type of giant virus called “Pithovirus” has been discovered in the frozen ground of extreme north-eastern Siberia by researchers from the Information Génomique et Structurale laboratory (CNRS/AMU), in association with teams from the Biologie à Grande Echelle laboratory (CEA/INSERM/Université Joseph Fourier), Génoscope (CEA/CNRS) and the Russian Academy of Sciences. Buried underground, this giant virus, which is harmless to humans and animals, has survived being frozen for more than 30,000 years. Although its size and amphora shape are reminiscent of Pandoravirus, analysis of its genome and replication mechanism proves that Pithovirus is very different. This work brings to three the number of distinct families of giant viruses. It is published on the website of the journal PNAS in the week of March 3, 2014. In the families Megaviridae (represented in particular by Mimivirus, discovered in 2003) and Pandoraviridae1, researchers thought they had classified the diversity of giant viruses (the only viruses visible under optical microscopy, since their diameter exceeds 0.5 microns). These viruses, which infect amoeba such as Acanthamoeba, contain a very large number of genes compared to common viruses (like influenza or AIDS, which only contain about ten genes). Their genome is about the same size or even larger than that of many bacteria. By studying a sample from the frozen ground of extreme north-eastern Siberia, in the Chukotka autonomous region, researchers were surprised to discover a new giant virus more than 30,000 years old (contemporaneous with the extinction of Neanderthal man), which they have named “Pithovirus sibericum”. -

Estimating Evolutionary Rates in Giant Viruses Using Ancient Genomes Sebastia´N Ducheˆne1 and Edward C
Virus Evolution, 2018, 4(1): vey006 doi: 10.1093/ve/vey006 Reflections Estimating evolutionary rates in giant viruses using ancient genomes Sebastia´n Ducheˆne1 and Edward C. Holmes2,*,† 1Department of Biochemistry and Molecular Biology, Bio21 Molecular Science and Biotechnology Institute, University of Melbourne, Parkville, VIC 3020, Australia and 2Marie Bashir Institute of Infectious Diseases and Biosecurity, Charles Perkins Centre, School of Life and Environmental Sciences and Sydney Medical School, University of Sydney, Sydney, NSW 2006, Australia *Corresponding author: E-mail: [email protected] †http://orcid.org/0000-0001-9596-3552 Abstract Pithovirus sibericum is a giant (610 Kpb) double-stranded DNA virus discovered in a purportedly 30,000-year-old permafrost sample. A closely related virus, Pithovirus massiliensis, was recently isolated from a sewer in southern France. An initial comparison of these two virus genomes assumed that P. sibericum was directly ancestral to P. massiliensis and gave a maximum evolutionary rate of 2.60 Â 10À5 nucleotide substitutions per site per year (subs/site/year). If correct, this would make pithoviruses among the fastest-evolving DNA viruses, with rates close to those seen in some RNA viruses. To help determine whether this unusually high rate is accurate we utilized the well-known negative association between evolution- ary rate and genome size in DNA microbes. This revealed that a more plausible rate estimate for Pithovirus evolution is 2.23 Â 10À6 subs/site/year, with even lower estimates obtained if evolutionary rates are assumed to be time-dependent. Hence, we estimate that Pithovirus has evolved at least an order of magnitude more slowly than previously suggested.