Evolution of Eukaryotic Cell Genomics, Phylogenomics & Biology
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Morphology, Phylogeny, and Diversity of Trichonympha (Parabasalia: Hypermastigida) of the Wood-Feeding Cockroach Cryptocercus Punctulatus
J. Eukaryot. Microbiol., 56(4), 2009 pp. 305–313 r 2009 The Author(s) Journal compilation r 2009 by the International Society of Protistologists DOI: 10.1111/j.1550-7408.2009.00406.x Morphology, Phylogeny, and Diversity of Trichonympha (Parabasalia: Hypermastigida) of the Wood-Feeding Cockroach Cryptocercus punctulatus KEVIN J. CARPENTER, LAWRENCE CHOW and PATRICK J. KEELING Canadian Institute for Advanced Research, Botany Department, University of British Columbia, University Boulevard, Vancouver, BC, Canada V6T 1Z4 ABSTRACT. Trichonympha is one of the most complex and visually striking of the hypermastigote parabasalids—a group of anaerobic flagellates found exclusively in hindguts of lower termites and the wood-feeding cockroach Cryptocercus—but it is one of only two genera common to both groups of insects. We investigated Trichonympha of Cryptocercus using light and electron microscopy (scanning and transmission), as well as molecular phylogeny, to gain a better understanding of its morphology, diversity, and evolution. Microscopy reveals numerous new features, such as previously undetected bacterial surface symbionts, adhesion of post-rostral flagella, and a dis- tinctive frilled operculum. We also sequenced small subunit rRNA gene from manually isolated species, and carried out an environmental polymerase chain reaction (PCR) survey of Trichonympha diversity, all of which strongly supports monophyly of Trichonympha from Cryptocercus to the exclusion of those sampled from termites. Bayesian and distance methods support a relationship between Tricho- nympha species from termites and Cryptocercus, although likelihood analysis allies the latter with Eucomonymphidae. A monophyletic Trichonympha is of great interest because recent evidence supports a sister relationship between Cryptocercus and termites, suggesting Trichonympha predates the Cryptocercus-termite divergence. -

Kleptoplast Photoacclimation State Modulates the Photobehaviour of the Solar-Powered Sea Slug Elysia Viridis
© 2018. Published by The Company of Biologists Ltd | Journal of Experimental Biology (2018) 221, jeb180463. doi:10.1242/jeb.180463 SHORT COMMUNICATION Kleptoplast photoacclimation state modulates the photobehaviour of the solar-powered sea slug Elysia viridis Paulo Cartaxana1, Luca Morelli1,2, Carla Quintaneiro1, Gonçalo Calado3, Ricardo Calado1 and Sónia Cruz1,* ABSTRACT light intensities, possibly as a strategy to prevent the premature loss Some sacoglossan sea slugs incorporate intracellular functional of kleptoplast photosynthetic function (Weaver and Clark, 1981; algal chloroplasts (kleptoplasty) for periods ranging from a few days Cruz et al., 2013). A distinguishable behaviour in response to light to several months. Whether this association modulates the has been recorded in Elysia timida: a change in the position of its photobehaviour of solar-powered sea slugs is unknown. In this lateral folds (parapodia) from a closed position to a spread, opened study, the long-term kleptoplast retention species Elysia viridis leaf-like posture under lower irradiance and the opposite behaviour showed avoidance of dark independently of light acclimation state. under high light levels (Rahat and Monselise, 1979; Jesus et al., In contrast, Placida dendritica, which shows non-functional retention 2010; Schmitt and Wägele, 2011). This behaviour in response to of kleptoplasts, showed no preference over dark, low or high light. light changes was only recorded for this species and has been assumed to be linked to long-term retention of kleptoplasts (Schmitt High light-acclimated (HLac) E. viridis showed a higher preference for and Wägele, 2011). high light than low light-acclimated (LLac) conspecifics. The position of the lateral folds (parapodia) was modulated by irradiance, with In this study, we determine whether the photobehaviour of increasing light levels leading to a closure of parapodia and protection the solar-powered sea slug Elysia viridis is linked to the of kleptoplasts from high light exposure. -

Frontiers in Zoology Biomed Central
Frontiers in Zoology BioMed Central Research Open Access Functional chloroplasts in metazoan cells - a unique evolutionary strategy in animal life Katharina Händeler*1, Yvonne P Grzymbowski1, Patrick J Krug2 and Heike Wägele1 Address: 1Zoologisches Forschungsmuseum Alexander Koenig, Adenauerallee 160, 53113 Bonn, Germany and 2Department of Biological Sciences, California State University, Los Angeles, California, 90032-8201, USA Email: Katharina Händeler* - [email protected]; Yvonne P Grzymbowski - [email protected]; Patrick J Krug - [email protected]; Heike Wägele - [email protected] * Corresponding author Published: 1 December 2009 Received: 26 June 2009 Accepted: 1 December 2009 Frontiers in Zoology 2009, 6:28 doi:10.1186/1742-9994-6-28 This article is available from: http://www.frontiersinzoology.com/content/6/1/28 © 2009 Händeler et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background: Among metazoans, retention of functional diet-derived chloroplasts (kleptoplasty) is known only from the sea slug taxon Sacoglossa (Gastropoda: Opisthobranchia). Intracellular maintenance of plastids in the slug's digestive epithelium has long attracted interest given its implications for understanding the evolution of endosymbiosis. However, photosynthetic ability varies widely among sacoglossans; some species have no plastid retention while others survive for months solely on photosynthesis. We present a molecular phylogenetic hypothesis for the Sacoglossa and a survey of kleptoplasty from representatives of all major clades. We sought to quantify variation in photosynthetic ability among lineages, identify phylogenetic origins of plastid retention, and assess whether kleptoplasty was a key character in the radiation of the Sacoglossa. -

Biology of Protists Video and DVD Guide. the BIOLOGY of PROTISTS Produced by Biomedia ASSOCIATES ©2003 -- Running Time 20 Minutes
Biology of Protists video and DVD guide. THE BIOLOGY OF PROTISTS Produced by BioMEDIA ASSOCIATES ©2003 -- Running time 20 minutes. Order Toll Free (877) 661-5355 or FAX (843) 470-0237 Or mail orders to eBioMEDIA, P.O. Box 1234, Beaufort, SC 29901–1234 (IMAGES IN THIS GUIDE ARE FROM THE VIDEO PROGRAM) Cosmerium, a green alga Arcella, a shelled ameoba The goal of this program is to show a representative sample of the great diversity of protists, and to show why they need a new classification reflecting our growing understanding of their long evolutionary history. The protists shown can be found in habitats such as: roadside puddles, park duck ponds, aquariums, birdbaths and in the gut of termites. We hope that these observations will encourage students to collect pond water samples and see for themselves this amazing hidden world. The live photography was accomplished using a variety microscope techniques, including dark field (good for showing the natural color of subjects) and the most advanced forms of differential interference contrast (DIC gives contrast to transparent structures that would be invisible in normal bright field microscopy). While these forms of imaging living protists are particularly revealing for some aspects of micro-organism biology, all of the organisms seen here can be studied successfully with student microscopes. IMPORTANT NOTE: This program packs a lot of information and a wealth of observational data into nine different modules. We recommend that the material be viewed module by module, stopping for discussion and replay as needed. “Discussion starters” for the various modules are provided in this guide. -

The Oxymonad Genome Displays Canonical Eukaryotic Complexity in the Absence of a Mitochondrion Anna Karnkowska,*,1,2 Sebastian C
The Oxymonad Genome Displays Canonical Eukaryotic Complexity in the Absence of a Mitochondrion Anna Karnkowska,*,1,2 Sebastian C. Treitli,1 Ondrej Brzon, 1 Lukas Novak,1 Vojtech Vacek,1 Petr Soukal,1 Lael D. Barlow,3 Emily K. Herman,3 Shweta V. Pipaliya,3 TomasPanek,4 David Zihala, 4 Romana Petrzelkova,4 Anzhelika Butenko,4 Laura Eme,5,6 Courtney W. Stairs,5,6 Andrew J. Roger,5 Marek Elias,4,7 Joel B. Dacks,3 and Vladimır Hampl*,1 1Department of Parasitology, BIOCEV, Faculty of Science, Charles University, Vestec, Czech Republic 2Department of Molecular Phylogenetics and Evolution, Faculty of Biology, Biological and Chemical Research Centre, University of Warsaw, Warsaw, Poland 3Division of Infectious Disease, Department of Medicine, University of Alberta, Edmonton, Canada 4Department of Biology and Ecology, Faculty of Science, University of Ostrava, Ostrava, Czech Republic Downloaded from https://academic.oup.com/mbe/article-abstract/36/10/2292/5525708 by guest on 13 January 2020 5Department of Biochemistry and Molecular Biology, Dalhousie University, Halifax, Canada 6Department of Cell and Molecular Biology, Uppsala University, Uppsala, Sweden 7Institute of Environmental Technologies, Faculty of Science, University of Ostrava, Ostrava, Czech Republic *Corresponding authors: E-mails: [email protected]; [email protected]. Associate editor: Fabia Ursula Battistuzzi Abstract The discovery that the protist Monocercomonoides exilis completely lacks mitochondria demonstrates that these organ- elles are not absolutely essential to eukaryotic cells. However, the degree to which the metabolism and cellular systems of this organism have adapted to the loss of mitochondria is unknown. Here, we report an extensive analysis of the M. -

A Resurgence in Field Research Is Essential to Better Understand
Received Date : 15-Mar-2013 Revised Date : 21-Oct-2013 Accepted Date : 29-Oct-2013 Article type : Symposium Article Heger et al. --- Importance of Field Protistology A Resurgence in Field Research is Essential to Better Understand the Diversity, Ecology, and Evolution of Microbial Eukaryotes Thierry J. Hegera, Virginia P. Edgcombb, Eunsoo Kimc, Julius Lukešd, Brian S. Leandera & Naoji Yubukia a The Departments of Botany and Zoology, Beaty Biodiversity Research Centre and Museum, University of British Columbia, Vancouver, British Columbia, V6T 1Z4, Canada b Woods Hole Oceanographic Institution, Geology and Geophysics Department, Woods Hole, Article MA 02543, USA c Division of Invertebrate Zoology, American Museum of Natural History, New York, NY, 10024, USA d Institute of Parasitology, Biology Centre, Czech Academy of Sciences, and Faculty of Science, University of South Bohemia, 37005 České Budějovice, Czech Republic Correspondence T. J. Heger and N. Yubuki, The Departments of Botany and Zoology, Beaty Biodiversity Research Centre and Museum, University of British Columbia, 6270 University Blvd., Vancouver, British Columbia, V6T 1Z4, Canada Telephone number: +1 604 822 4892; FAX number: +1 604 822 6089; emails: [email protected] and [email protected] ABSTRACT The discovery and characterization of protist communities from diverse environments are crucial for understanding the overall evolutionary history of life on earth. However, major questions about the diversity, ecology, and evolutionary history of protists remain unanswered, -
Prokaryotic and Eukaryotic Organisms Pdf
Prokaryotic and eukaryotic organisms pdf Continue There are two types of cells: prokaryotic and eukaryotic. In this section, we will examine the similarities and differences between the two types. The objectives of the training to identify features common to all cells contrast the composition and size of prokaryotic and eukaryotic cells fall into one of two broad categories: prokaryotic and eukaryotic. Single-celled organisms of the domains Of Bacteria and Archaea are classified as prokaryotes (pro - before; carion - core). Animal cells, plant cells, fungi and proteanists are eukaryotes (eu - truth). Components of prokaryotic cells All cells have four common components: (1) plasma membrane, external coating separating the inner part of the cell from the environment; (2) cytoplasm, consisting of a jelly-like area inside the cell in which other cellular components are located; (3) DNA, the genetic material of the cell; and (4) ribosomes, particles that synthesize proteins. However, prokaryotes differ from eukaryotic cells in several ways. Figure 1. This image shows the generalized structure of the prokaryotic cell. A prokaryotic cell is a simple single-celled organism that lacks a nucleus or any other membrane organella. Soon we will see that this is significantly different in eukaryotes. Prokaryotic DNA is found in the central part of the cell: a darkened area called a nucleoid (Figure 1). Unlike archaea and eukaryote, bacteria have a cell wall of peptidoglycan consisting of sugars and amino acids, and many of them have a polysaccharide capsule (Figure 1). The cell wall acts as an additional layer of protection, helps the cell maintain its shape and prevents dehydration. -
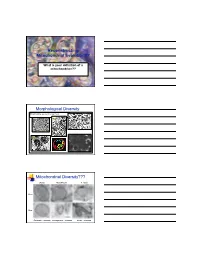
Reconstructing Mitochondrial Evolution?? Morphological Diversity Mitochondrial Diversity???
Reconstructing Mitochondrial Evolution?? What is your definition of a mitochondrion?? Morphological Diversity Mitochondria as we all know them: Suprarenal gland Liver cell Plasma cell Adrenal cortex Mitochondrial Diversity??? Chicken Neocallimastix T. foetus 100 nm 50 nm Entamoeba - mitosome microsporidian - mitosome Giardia - mitosome Reconstructing Evolution Mitochondrial evolution well established endosymbiotic theory α-proteobacterium - Rickettsia prowazekii Hydrogenosomal evolution No DNA NOW 2 examples Nyctotherus and Blastocystis (MLO) Several proteins similar to mitochondria Mitosome evolution No DNA Few proteins identified similar to mitochondria Origins via Endosymbiosis Current dogma - mitochondria and related Aerobic α-proteobacterium prokaryote gave rise to present day mitochondria. organelles arose just once in evolution Are hydrogenosomes and mitosomes of anaerobic protists derived from the same proto-mitochondrion? Evidence for: accumulating evidence for several proteins that are currently found in mitochondria - Proteins of Fe-S cluster formation. Scenario A Common ancestor facultative Scenario B organism? degenerate mitochondrion invoke lateral gene transfer from anaerobic prokaryotes Hypotheses for Mito Acquisition Mitochondrion-related Organelles 2 way to interpret this summary - can you think of both? Groups with mitochondrial homologues Hjort et al Phil. Trans. R. Soc. B 2010 365, 713-727 Origin of Hydrogenosomes (Mitosomes?) 1. a)Conversion of Mitochondria HYD b)Common ancestor with mitochondria ? HYD 2. Independent Origin α-proteobacterium ? - anaerobic bacterium HYD Organelles - origins and biogenesis Approaches: (1) Conduct phylogenetic analyses of similar proteins Hsp70 Fd Hsp60 Isc subunits (2) Examine protein targeting to the organelle matrix protein targeting membrane protein targeting (3) Characterize membrane/translocation components These components could have evolved as the endosymbiont was converted to organelle. Reveals evolutionary history. -

Novel Lineages of Oxymonad Flagellates from the Termite Porotermes Adamsoni (Stolotermitidae): the Genera Oxynympha and Termitim
Protist, Vol. 170, 125683, December 2019 http://www.elsevier.de/protis Published online date 21 October 2019 ORIGINAL PAPER Novel Lineages of Oxymonad Flagellates from the Termite Porotermes adamsoni (Stolotermitidae): the Genera Oxynympha and Termitimonas a,1 b a c b,1 Renate Radek , Katja Meuser , Samet Altinay , Nathan Lo , and Andreas Brune a Evolutionary Biology, Institute for Biology/Zoology, Freie Universität Berlin, 14195 Berlin, Germany b Research Group Insect Gut Microbiology and Symbiosis, Max Planck Institute for Terrestrial Microbiology, 35043 Marburg, Germany c School of Life and Environmental Sciences, The University of Sydney, Sydney, NSW 2006, Australia Submitted January 21, 2019; Accepted October 9, 2019 Monitoring Editor: Alastair Simpson The symbiotic gut flagellates of lower termites form host-specific consortia composed of Parabasalia and Oxymonadida. The analysis of their coevolution with termites is hampered by a lack of informa- tion, particularly on the flagellates colonizing the basal host lineages. To date, there are no reports on the presence of oxymonads in termites of the family Stolotermitidae. We discovered three novel, deep-branching lineages of oxymonads in a member of this family, the damp-wood termite Porotermes adamsoni. One tiny species (6–10 m), Termitimonas travisi, morphologically resembles members of the genus Monocercomonoides, but its SSU rRNA genes are highly dissimilar to recently published sequences of Polymastigidae from cockroaches and vertebrates. A second small species (9–13 m), Oxynympha loricata, has a slight phylogenetic affinity to members of the Saccinobaculidae, which are found exclusively in wood-feeding cockroaches of the genus Cryptocercus, the closest relatives of termites, but shows a combination of morphological features that is unprecedented in any oxymonad family. -

Parasitology Mgr. Anna Novák Vanclová Evolution of Euglenid Plastid Proteome
Charles University, Faculty of Science Univerzita Karlova, Přírodovědecká fakulta Study program: Parasitology Studijní program: Parazitologie Mgr. Anna Novák Vanclová Evolution of euglenid plastid proteome Evoluce proteomu plastidu euglenidů Summary of the doctoral thesis Thesis supervisor: doc. Vladimír Hampl, Ph.D. Prague, 2019 ABSTRACT Endosymbiotic gain and transfer of plastids is a widespread evolutionary phenomenon and a major driving force of eukaryotic evolution. The integration of a new organelle is accompanied by changes in its structure, gene content, molecular mechanisms for biogenesis and transport, and re-wiring of the host and organelle metabolic pathways. To understand the course and underlying mechanisms of plastid evolution, it is important to study these processes in variety of secondary algae and notice their differences and similarities. Euglenophytes gained their plastids from green eukaryotic algae after a long history of heterotrophic lifestyle. In my thesis, I participated in analyses of newly generated sequence datasets: transcriptomes of Euglena gracilis and Euglena longa and mass spectrometry-determined proteome of E. gracilis plastid with especial regard to the potential novelties associated with plastid gain and incorporation. In the resulting publications we particularly focus on plastid protein import machinery and targeting signals and report extremely reduced TIC and completely absent TOC in euglenophyte plastid. Using the proteomic dataset, we predict potential novel plastid protein translocases recruited from ER/Golgi and re-analyze plastid signal domains, characterizing previously overlooked features. Protein inventory of E. gracilis plastid suggests complex, in some cases redundant metabolic capacity. Chlorophyll recycling is one of the sources of phytol for reactions not connected to MEP/DOXP pathway. -

Mitosomes in Entamoeba Histolytica Contain a Sulfate Activation Pathway
Mitosomes in Entamoeba histolytica contain a sulfate activation pathway Fumika Mi-ichi, Mohammad Abu Yousuf, Kumiko Nakada-Tsukui, and Tomoyoshi Nozaki1 Department of Parasitology, National Institute of Infectious Diseases, Tokyo 162-8640, Japan Edited by Andrew Roger, Dalhousie University, Halifax, NS, Canada, and accepted by the Editorial Board October 19, 2009 (received for review June 25, 2009) Hydrogenosomes and mitosomes are mitochondrion-related or- and M. balamuthi lack the ISC system, and instead possess the ganelles in anaerobic/microaerophilic eukaryotes with highly re- nitrogen fixation (NIF) system, which is most likely derived from an duced and divergent functions. The full diversity of their content ancestral nitrogen fixing -proteobacterium by lateral gene transfer and function, however, has not been fully determined. To under- (22, 24). Only 5 proteins have been demonstrated in E. histolytica stand the central role of mitosomes in Entamoeba histolytica,a mitosomes: Cpn60 (8–10, 12), Cpn10 (13), mitochondrial Hsp70 parasitic protozoon that causes amoebic dysentery and liver ab- (11, 15), pyridine nucleotide transhydrogenase (PNT) (2, 8), and scesses, we examined the proteomic profile of purified mitosomes. mitochondria carrier family (MCF, ADP/ATP transporter) (14), Using 2 discontinuous Percoll gradient centrifugation and MS and the central role of mitosomes in E. histolytica remains unknown. analysis, we identified 95 putative mitosomal proteins. Immuno- Analysis of the genome of E. histolytica has not revealed any fluorescence assay showed that 3 proteins involved in sulfate additional information regarding the function of mitosomes and activation, ATP sulfurylase, APS kinase, and inorganic pyrophos- thus, a proteomic analysis of mitosomes seems to be the best phatase, as well as sodium/sulfate symporter, involved in sulfate approach to understand its structure and function (1, 2). -

BMC Evolutionary Biology Biomed Central
BMC Evolutionary Biology BioMed Central Research article Open Access Complex distribution of EFL and EF-1α proteins in the green algal lineage Geoffrey P Noble, Matthew B Rogers and Patrick J Keeling* Address: Canadian Institute for Advanced Research, Department of Botany, University of British Columbia, 3529-6270 University Boulevard, Vancouver, BC V6T 1Z4, Canada Email: Geoffrey P Noble - [email protected]; Matthew B Rogers - [email protected]; Patrick J Keeling* - [email protected] * Corresponding author Published: 23 May 2007 Received: 5 February 2007 Accepted: 23 May 2007 BMC Evolutionary Biology 2007, 7:82 doi:10.1186/1471-2148-7-82 This article is available from: http://www.biomedcentral.com/1471-2148/7/82 © 2007 Noble et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background: EFL (or elongation factor-like) is a member of the translation superfamily of GTPase proteins. It is restricted to eukaryotes, where it is found in a punctate distribution that is almost mutually exclusive with elongation factor-1 alpha (EF-1α). EF-1α is a core translation factor previously thought to be essential in eukaryotes, so its relationship to EFL has prompted the suggestion that EFL has spread by horizontal or lateral gene transfer (HGT or LGT) and replaced EF-1α multiple times. Among green algae, trebouxiophyceans and chlorophyceans have EFL, but the ulvophycean Acetabularia and the sister group to green algae, land plants, have EF-1α.