Openfoam Postprocessing and Advanced Running Options
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Arxiv:1911.09220V2 [Cs.MS] 13 Jul 2020
MFEM: A MODULAR FINITE ELEMENT METHODS LIBRARY ROBERT ANDERSON, JULIAN ANDREJ, ANDREW BARKER, JAMIE BRAMWELL, JEAN- SYLVAIN CAMIER, JAKUB CERVENY, VESELIN DOBREV, YOHANN DUDOUIT, AARON FISHER, TZANIO KOLEV, WILL PAZNER, MARK STOWELL, VLADIMIR TOMOV Lawrence Livermore National Laboratory, Livermore, USA IDO AKKERMAN Delft University of Technology, Netherlands JOHANN DAHM IBM Research { Almaden, Almaden, USA DAVID MEDINA Occalytics, LLC, Houston, USA STEFANO ZAMPINI King Abdullah University of Science and Technology, Thuwal, Saudi Arabia Abstract. MFEM is an open-source, lightweight, flexible and scalable C++ library for modular finite element methods that features arbitrary high-order finite element meshes and spaces, support for a wide variety of dis- cretization approaches and emphasis on usability, portability, and high-performance computing efficiency. MFEM's goal is to provide application scientists with access to cutting-edge algorithms for high-order finite element mesh- ing, discretizations and linear solvers, while enabling researchers to quickly and easily develop and test new algorithms in very general, fully unstructured, high-order, parallel and GPU-accelerated settings. In this paper we describe the underlying algorithms and finite element abstractions provided by MFEM, discuss the software implementation, and illustrate various applications of the library. arXiv:1911.09220v2 [cs.MS] 13 Jul 2020 1. Introduction The Finite Element Method (FEM) is a powerful discretization technique that uses general unstructured grids to approximate the solutions of many partial differential equations (PDEs). It has been exhaustively studied, both theoretically and in practice, in the past several decades [1, 2, 3, 4, 5, 6, 7, 8]. MFEM is an open-source, lightweight, modular and scalable software library for finite elements, featuring arbitrary high-order finite element meshes and spaces, support for a wide variety of discretization approaches and emphasis on usability, portability, and high-performance computing (HPC) efficiency [9]. -

Elmer Finite Element Software for Multiphysical Problems
Elmer finite element software for multiphysical problems Peter Råback ElmerTeam CSC – IT Center for Science 10/04/2014 PRACE Spring School, 15-17 April, 2014 1 Elmer track in PRACE spring school • Wed 16th, 10.45-12.15=1.5 h Introduction to Elmer finite element software • Wed 16th, 13.30-15.00=1.5 h Hands-on session using ElmerGUI • Wed 16th, 15.15-16.45=1.5 h OpenLab • Thu 17th, 10.45-12.15=1.5 h Advanced use of Elmer • Thu 17th, 13.30-14.30=1 h Hands-on session on advanced features and parallel computation • Thu 17th, 14.45-15.45=1h Hands-on & improviced 10/04/2014 2 Elmer finite element software for multiphysical problems Figures by Esko Järvinen, Mikko Lyly, Peter Råback, Timo Veijola (TKK) & Thomas Zwinger Short history of Elmer 1995 Elmer development was started as part of a national CFD program – Collaboration of CSC, TKK, VTT, JyU, and Okmetic Ltd. 2000 After the initial phase the development driven by number of application projects – MEMS, Microfluidics, Acoustics, Crystal Growth, Hemodynamics, Glaciology, … 2005 Elmer published under GPL-license 2007 Elmer version control put under sourceforge.net – Resulted to a rapid increase in the number of users 2010 Elmer became one of the central codes in PRACE project 2012 ElmerSolver library published under LGPL – More freedom for serious developers Elmer in numbers ~350,000 lines of code (~2/3 in Fortran, 1/3 in C/C++) ~500 code commits yearly ~280 consistency tests in 3/2014 ~730 pages of documentation in LaTeX ~60 people participated on Elmer courses in 2012 9 Elmer related visits -

Overview of Elmer
Overview of Elmer Peter Råback and Mika Malinen CSC – IT Center for Science October 19, 2012 1 2 Copyright This document is licensed under the Creative Commons Attribution-No Derivative Works 3.0 License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nd/3.0/. 1 Introduction This exposition gives an overview of the Elmer software package. General information on the capabilities of the software, its usage, and how the material of the package is organized is presented. More detailed information is given in the other Elmer manuals, the scopes of which are described in this document. What is Elmer Elmer is a finite element software package for the solution of partial differential equations. Elmer can deal with a great number of different equations, which may be coupled in a generic manner making Elmer a versatile tool for multiphysical simulations. As an open source software, Elmer also gives the user the means to modify the existing solution procedures and to develop new solvers for equations of interest to the user. History of Elmer The development of Elmer was started in 1995 as part of a national CFD technology program funded by the Finnish funding agency for technology and innovation, Tekes. The original development consortia included partners from CSC – IT Center for Science (formely known as CSC – Scientific Computing). Helsinki University of Technology TKK, VTT Technical Research Centre of Finland, University of Jyväskylä, and Okmetic Ltd. CSC is a governmental non-profit company fully owned by the Ministry of Education. After the five years initial project ended the development has been continued by CSC in different application fields. -

Customizing Paraview
Customizing ParaView Utkarsh A. Ayachit∗ David E. DeMarle† Kitware Inc. ABSTRACT VTK developers must overcome in order to extend ParaView. For- ParaView is an Open Source Visualization Application that scales, tunately the ServerManager includes both Module [2] and server via data parallel processing, to massive problem sizes. The nature side Plugin [3] facilities. The two facilities differ in that Modules of Open Source software means that ParaView has always been well are statically linked into ParaView at compile time, whereas Plug- suited to customization. However having access to the source code ins are dynamically linked into ParaView at run time. Either facility does not imply that it is trivial to extend or reuse the application. An standardizes, through CMake[6] configuration macros, the process ongoing goal of the ParaView project is to make the code simple to by which new types of VTK objects can be added to ParaView. Plu- extend, customize, and change arbitrarily. gin and Module macros turn the build environment definition into In this paper we present the ways by which the application can be a simple procedure call. A standard list of arguments is populated modified to date. We then describe in more detail the latest efforts with information such as the names of the C++ files for the new rou- to simplify the task of reusing the most visible part of ParaView, the tines, and the macro creates the correct platform independent build client GUI application. These efforts include a new CMake config- environment to make a library that is compatible with the applica- uration macro that makes it simple to assemble the major functional tion. -

Usr/Bin/Cmake # You Can Edit This File to Change Values Found and Used by Cmake
# This is the CMakeCache file. # For build in directory: /home/build2 # It was generated by CMake: /usr/bin/cmake # You can edit this file to change values found and used by cmake. # If you do not want to change any of the values, simply exit the editor. # If you do want to change a value, simply edit, save, and exit the editor. # The syntax for the file is as follows: # KEY:TYPE=VALUE # KEY is the name of a variable in the cache. # TYPE is a hint to GUIs for the type of VALUE, DO NOT EDIT TYPE!. # VALUE is the current value for the KEY. ######################## # EXTERNAL cache entries ######################## //Build shared libraries (so/dylib/dll). BUILD_SHARED_LIBS:BOOL=ON //Build testing BUILD_TESTING:BOOL=OFF //Path to a program. BZRCOMMAND:FILEPATH=BZRCOMMAND-NOTFOUND //Path to a program. CMAKE_AR:FILEPATH=/usr/bin/ar //The build mode CMAKE_BUILD_TYPE:STRING=Release //The build type for the paraview project. CMAKE_BUILD_TYPE_paraview:STRING=Release //Enable/Disable color output during build. CMAKE_COLOR_MAKEFILE:BOOL=ON //CXX compiler CMAKE_CXX_COMPILER:FILEPATH=/usr/bin/c++ //A wrapper around 'ar' adding the appropriate '--plugin' option // for the GCC compiler CMAKE_CXX_COMPILER_AR:FILEPATH=/usr/bin/gcc-ar //A wrapper around 'ranlib' adding the appropriate '--plugin' option // for the GCC compiler CMAKE_CXX_COMPILER_RANLIB:FILEPATH=/usr/bin/gcc-ranlib //Flags used by the CXX compiler during all build types. CMAKE_CXX_FLAGS:STRING= //Flags used by the CXX compiler during DEBUG builds. CMAKE_CXX_FLAGS_DEBUG:STRING=-g //Flags used by the CXX compiler during MINSIZEREL builds. CMAKE_CXX_FLAGS_MINSIZEREL:STRING=-Os -DNDEBUG //Flags used by the CXX compiler during RELEASE builds. CMAKE_CXX_FLAGS_RELEASE:STRING=-O2 -DNDEBUG //Flags used by the CXX compiler during RELWITHDEBINFO builds. -

Introduction to Visualization with VTK and Paraview
Outline Introduction Visualization with ParaView Showcases Resources and further reading Introduction to Visualization with VTK and ParaView R. Sungkorn and J. Derksen Department of Chemical and Materials Engineering University of Alberta Canada August 24, 2011 / LBM Workshop Outline Introduction Visualization with ParaView Showcases Resources and further reading 1 Introduction Simulation Visualization Visualization Pipeline Data Structure Data Format ParaView 2 Visualization with ParaView ParaView Interface Loading Data View Controls Structure Filters Save data and animation 3 Showcases 4 Resources and further reading Outline Introduction Visualization with ParaView Showcases Resources and further reading Simulation Visualization Simulation visualization referred to a computing method in which a geometric representation is used to gain understanding and insight into numeric data generated by numerical simulation. The data is usually placed into a reference coordinate system to create and extract quantities/qualities of interest. Figure 1: Streamlines of flow pass a cylinder (image courtesy of Kitware Inc). Outline Introduction Visualization with ParaView Showcases Resources and further reading Visualization Pipeline The goal of visualization pipeline is to create geometrically constructed images from numeric data. The process can be described step-wise as: Data analysis: preparing data for visualization Filtering: specifying data portion to be visualized Mapping: transforming filtered data into geometrical primitive (e.g. points, lines) with attributes (e.g. color, size) Rendering: generating image from geometric data Figure 2: Visualization pipeline (image courtesy of www.infovis-wiki.net). Outline Introduction Visualization with ParaView Showcases Resources and further reading Data Structure Data structure is a way of exporting and organizing simulated data. It also defines applicability of some visualization techniques (e.g. -

Pipenightdreams Osgcal-Doc Mumudvb Mpg123-Alsa Tbb
pipenightdreams osgcal-doc mumudvb mpg123-alsa tbb-examples libgammu4-dbg gcc-4.1-doc snort-rules-default davical cutmp3 libevolution5.0-cil aspell-am python-gobject-doc openoffice.org-l10n-mn libc6-xen xserver-xorg trophy-data t38modem pioneers-console libnb-platform10-java libgtkglext1-ruby libboost-wave1.39-dev drgenius bfbtester libchromexvmcpro1 isdnutils-xtools ubuntuone-client openoffice.org2-math openoffice.org-l10n-lt lsb-cxx-ia32 kdeartwork-emoticons-kde4 wmpuzzle trafshow python-plplot lx-gdb link-monitor-applet libscm-dev liblog-agent-logger-perl libccrtp-doc libclass-throwable-perl kde-i18n-csb jack-jconv hamradio-menus coinor-libvol-doc msx-emulator bitbake nabi language-pack-gnome-zh libpaperg popularity-contest xracer-tools xfont-nexus opendrim-lmp-baseserver libvorbisfile-ruby liblinebreak-doc libgfcui-2.0-0c2a-dbg libblacs-mpi-dev dict-freedict-spa-eng blender-ogrexml aspell-da x11-apps openoffice.org-l10n-lv openoffice.org-l10n-nl pnmtopng libodbcinstq1 libhsqldb-java-doc libmono-addins-gui0.2-cil sg3-utils linux-backports-modules-alsa-2.6.31-19-generic yorick-yeti-gsl python-pymssql plasma-widget-cpuload mcpp gpsim-lcd cl-csv libhtml-clean-perl asterisk-dbg apt-dater-dbg libgnome-mag1-dev language-pack-gnome-yo python-crypto svn-autoreleasedeb sugar-terminal-activity mii-diag maria-doc libplexus-component-api-java-doc libhugs-hgl-bundled libchipcard-libgwenhywfar47-plugins libghc6-random-dev freefem3d ezmlm cakephp-scripts aspell-ar ara-byte not+sparc openoffice.org-l10n-nn linux-backports-modules-karmic-generic-pae -

Paraview and Python Paraview Offers Rich Scripting Support Through Python
Version 4.0 Contents Articles Introduction 1 About Paraview 1 Loading Data 9 Data Ingestion 9 Understanding Data 13 VTK Data Model 13 Information Panel 23 Statistics Inspector 27 Memory Inspector 28 Multi-block Inspector 32 Displaying Data 35 Views, Representations and Color Mapping 35 Filtering Data 70 Rationale 70 Filter Parameters 70 The Pipeline 72 Filter Categories 76 Best Practices 79 Custom Filters aka Macro Filters 82 Quantative Analysis 85 Drilling Down 85 Python Programmable Filter 85 Calculator 92 Python Calculator 94 Spreadsheet View 99 Selection 102 Querying for Data 111 Histogram 115 Plotting and Probing Data 116 Saving Data 118 Saving Data 118 Exporting Scenes 121 3D Widgets 123 Manipulating data in the 3D view 123 Annotation 130 Annotation 130 Animation 141 Animation View 141 Comparative Visualization 149 Comparative Views 149 Remote and Parallel Large Data Visualization 156 Parallel ParaView 156 Starting the Server(s) 159 Connecting to the Server 164 Distributing/Obtaining Server Connection Configurations 168 Parallel Rendering and Large Displays 172 About Parallel Rendering 172 Parallel Rendering 172 Tile Display Walls 178 CAVE Displays 180 Scripted Control 188 Interpreted ParaView 188 Python Scripting 188 Tools for Python Scripting 210 Batch Processing 212 In-Situ/CoProcessing 217 CoProcessing 217 C++ CoProcessing example 229 Python CoProcessing Example 236 Plugins 242 What are Plugins? 242 Included Plugins 243 Loading Plugins 245 Appendix 248 Command Line Arguments 248 Application Settings 255 List of Readers 264 List -

Post-Processing with Paraview
Post-processing with Paraview R. Ponzini, CINECA -SCAI Post-processing with Paraview: Overall Program Post-processing with Paraview I (ParaView GUI and Filters) Post-processing with Paraview II (ParaView scripting with hands-on) Post-processing with Paraview III (ParaView for large data visualization) OUTLINE PART A PART B • Filters • What is Paraview • Vectors visualization • The GUI • Sources • Streamlines • Loading Data • Plotting over line • Text annotation • Select data • Views • Create a custom filter management • Animations • Save figures • Time dependent data What is Paraview ParaView is an open-source application for visualizing 2D/3D data. To date, ParaView has been demonstrated to process billions of unstructured cells and to process over a trillion structured cells. ParaView's parallel framework has run on over 100,000 processing cores. ParaView's key features are: • An open-source, scalable, multi-platform visualization application. • Support for distributed computation models to process large data sets. • An open, extensible, and intuitive user interface. • An extensible, modular architecture based on open standards. • A flexible BSD 3-clause license. • Commercial maintenance and support. PARAVIEW: a standard de-facto ParaView is used by many academic, government, and commercial institutions all over the world. ParaView is downloaded roughly 100,000 times every year. ParaView also won the HPCwire Readers' Choice Award and HPCwire Editors' Choice Award for Best HPC Visualization Product or Technology. Obtaining Paraview & Official Resources • Main website: http://www.paraview.org/ • Download page: http://www.paraview.org/paraview/resources/software.php • Resources (video): http://www.paraview.org/paraview/resources/webinars.html • Resources (wiki): http://www.paraview.org/Wiki/ParaView The big picture The application most people associate with ParaView is really just a small client application built on top of a tall stack of libraries that provide ParaView with its functionality. -
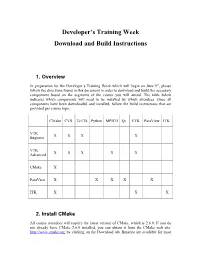
Developer's Training Week Download and Build Instructions
Developer’s Training Week Download and Build Instructions 1. Overview In preparation for the Developer’s Training Week which will begin on June 9th, please follow the directions found in this document in order to download and build the necessary components based on the segments of the course you will attend. The table below indicates which components will need to be installed by which attendees. Once all components have been downloaded and installed, follow the build instructions that are provided per course topic. CMake CVS Tcl/Tk Python MPICH Qt VTK ParaView ITK VTK X X X X Beginner VTK X X X X X Advanced CMake X ParaView X X X X X ITK X X X 2. Install CMake All course attendees will require the latest version of CMake, which is 2.6.0. If you do not already have CMake 2.6.0 installed, you can obtain it from the CMake web site: http://www.cmake.org by clicking on the Download tab. Binaries are available for most platforms, so download and install the binary appropriate for your particular operating system. If binaries are not available for your platform, download the CMake source code, and follow the instructions contained in the download. In addition, if binaries are not available for your platform, please let us know what operating system you will be using during the course. 3. Install CVS Attendees for the VTK sessions will need to have CVS installed in order to obtain the source code to VTK 5.2. You can obtain CVS through http://www.nongnu.org/cvs/. -

Authors: Manuel Carmona José María Gómez José Bosch Manel López Óscar Ruiz
Authors: Manuel Carmona José María Gómez José Bosch Manel López Óscar Ruiz 1/53 November/2019 Barcelona, Spain. This work is licensed under a Creative Commons Attribution-NonCommercial- NoDerivs 3.0 Unported License http://creativecommons.org/licenses/by-nc-nd/3.0 2/53 Table of contents I. Introduction to Elmer .................................................................................................................... 5 II. Elmer GUI ..................................................................................................................................... 6 III. Elmer commands ........................................................................................................................ 10 III.1. Header section ...................................................................................................................... 11 III.2. Constants section ................................................................................................................. 12 III.3. Simulation section ................................................................................................................ 12 III.4. Body section ........................................................................................................................ 13 III.5. Material section .................................................................................................................... 14 III.6. Body Force section ............................................................................................................. -

Scientific Visualization with Paraview
Scientific Visualization with ParaView Geilo Winter School 2016 Andrea Brambilla (GEXCON AS, Bergen) Outline • Part 1 (Monday) • Fundamentals • Data Filtering • Part 2 (Tuesday) • Time Dependent Data • Selection & Linked Views • Part 3 (Thursday) • Python scripting Requirements • Install ParaView 4.4 • http://www.paraview.org Download • Get example material • http://www.paraview.org/Wiki/The_ParaView_Tutorial • Additional data on the thumb drives • On the thumb drives: • ParaView installers • Tutorial data • These slides, VTK Documentation (useful for python) Introduction What is ParaView? • An open-source, scalable, multi-platform visualization application. • Support for distributed computation models to process large data sets. • An open, flexible, and intuitive user interface. • An extensible, modular architecture based on open standards. • Large user community, both public and private sector. • A flexible BSD 3 Clause license. Renato N. Elias, NACAD/COPPE/UFRJ, Rio de Janerio, Brazil Jerry Clarke, US Army Research Laboratory Swiss National Supercomputing Centre Bill Daughton, LANL Swiss National Supercomputing Centre ParaView Application Architecture ParaView Client pvpython ParaWeb Catalyst Custom App UI (Qt Widgets, Python Wrappings) ParaView Server VTK OpenGL MPI IceT Etc. ParaView Development • Started in 2000 as collaborative effort between Los Alamos National Laboratories and Kitware Inc. (lead by James Ahrens) • ParaView 0.6 released October 2002 • September 2005: collaborative effort between Sandia National Laboratories, Kitware