Estimating Daily Primary Production and Nighttime Respiration in Estuaries by an in Situ Carbon Method
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Net Ecosystem Production: a Comprehensive Measure of Net Carbon Accumulation by Ecosystems
Ecological Applications, 12(4), 2002, pp. 937±947 q 2002 by the Ecological Society of America NET ECOSYSTEM PRODUCTION: A COMPREHENSIVE MEASURE OF NET CARBON ACCUMULATION BY ECOSYSTEMS J. T. RANDERSON,1,6 F. S . C HAPIN, III,2 J. W. HARDEN,3 J. C. NEFF,4 AND M. E. HARMON5 1Divisions of Engineering and Applied Science and Geological and Planetary Sciences, California Institute of Technology, Mail Stop 100-23, Pasadena, California 91125 USA 2Institute for Arctic Biology, 311 Irvine 1 Building, University of Alaska, Fairbanks, Fairbanks, Alaska, 99775 USA 3U.S. Geological Survey, Mail Stop 962, 345 Middle®eld Road, Menlo Park, California 94025 USA 4U.S. Geological Survey, Mail Stop 980, Denver Federal Center, Denver, Colorado 80225 USA 5Department of Forest Science, Oregon State University, Corvallis, Oregon 97331-57521 USA Abstract. The conceptual framework used by ecologists and biogeochemists must allow for accurate and clearly de®ned comparisons of carbon ¯uxes made with disparate tech- niques across a spectrum of temporal and spatial scales. Consistent with usage over the past four decades, we de®ne ``net ecosystem production'' (NEP) as the net carbon accu- mulation by ecosystems. Past use of this term has been ambiguous, because it has been used conceptually as a measure of carbon accumulation by ecosystems, but it has often been calculated considering only the balance between gross primary production (GPP) and ecosystem respiration. This calculation ignores other carbon ¯uxes from ecosystems (e.g., leaching of dissolved carbon and losses associated with disturbance). To avoid conceptual ambiguities, we argue that NEP be de®ned, as in the past, as the net carbon accumulation by ecosystems and that it explicitly incorporate all the carbon ¯uxes from an ecosystem, including autotrophic respiration, heterotrophic respiration, losses associated with distur- bance, dissolved and particulate carbon losses, volatile organic compound emissions, and lateral transfers among ecosystems. -

Spatial and Temporal Variation in Sediment-Associated Microbial Respiration in Oil Sands Mine-Affected Wetlands of North-Eastern Alberta, Canada
University of Windsor Scholarship at UWindsor Electronic Theses and Dissertations Theses, Dissertations, and Major Papers 2010 Spatial and temporal variation in sediment-associated microbial respiration in oil sands mine-affected wetlands of north-eastern Alberta, Canada. Jesse Gardner Costa University of Windsor Follow this and additional works at: https://scholar.uwindsor.ca/etd Recommended Citation Gardner Costa, Jesse, "Spatial and temporal variation in sediment-associated microbial respiration in oil sands mine-affected wetlands of north-eastern Alberta, Canada." (2010). Electronic Theses and Dissertations. 282. https://scholar.uwindsor.ca/etd/282 This online database contains the full-text of PhD dissertations and Masters’ theses of University of Windsor students from 1954 forward. These documents are made available for personal study and research purposes only, in accordance with the Canadian Copyright Act and the Creative Commons license—CC BY-NC-ND (Attribution, Non-Commercial, No Derivative Works). Under this license, works must always be attributed to the copyright holder (original author), cannot be used for any commercial purposes, and may not be altered. Any other use would require the permission of the copyright holder. Students may inquire about withdrawing their dissertation and/or thesis from this database. For additional inquiries, please contact the repository administrator via email ([email protected]) or by telephone at 519-253-3000ext. 3208. Spatial and temporal variation in sediment-associated microbial respiration in oil sands mine-affected wetlands of north-eastern Alberta, Canada. by Jesse M. Gardner Costa A Thesis Submitted to the Faculty of Graduate Studies through the Department of Biological Sciences in partial fulfillment of the requirements for the Degree of Master of Science at the University of Windsor Windsor, Ontario, Canada 2010 © 2010 Jesse M. -

Analytically Tractable Climate-Carbon Cycle Feedbacks Under 21St Century Anthropogenic Forcing Steven J
Analytically tractable climate-carbon cycle feedbacks under 21st century anthropogenic forcing Steven J. Lade1,2,3, Jonathan F. Donges1,4, Ingo Fetzer1,3, John M. Anderies5, Christian Beer3,6, Sarah E. Cornell1, Thomas Gasser7, Jon Norberg1, Katherine Richardson8, Johan Rockström1, and Will Steffen1,2 1Stockholm Resilience Centre, Stockholm University, Stockholm, Sweden 2Fenner School of Environment and Society, The Australian National University, Canberra, Australian Capital Territory, Australia 3Bolin Centre for Climate Research, Stockholm University, Stockholm, Sweden 4Potsdam Institute for Climate Impact Research, Potsdam, Germany 5School of Sustainability and School of Human Evolution and Social Change, Arizona State University, Tempe, Arizona, USA 6Department of Environmental Science and Analytical Chemistry (ACES), Stockholm University, Stockholm, Sweden 7International Institute for Applied Systems Analysis, Laxenburg, Austria 8Center for Macroecology, Evolution, and Climate, University of Copenhagen, Natural History Museum of Denmark, Copenhagen, Denmark Correspondence to: Steven Lade ([email protected]) Abstract. Changes to climate-carbon cycle feedbacks may significantly affect the Earth System’s response to greenhouse gas emissions. These feedbacks are usually analysed from numerical output of complex and arguably opaque Earth System Models (ESMs). Here, we construct a stylized global climate-carbon cycle model, test its output against complex comprehensive ESMs, and investigate the strengths of its climate-carbon cycle feedbacks -

21St-Century Modeled Permafrost Carbon Emissions Accelerated by Abrupt Thaw Beneath Lakes
ARTICLE DOI: 10.1038/s41467-018-05738-9 OPEN 21st-century modeled permafrost carbon emissions accelerated by abrupt thaw beneath lakes Katey Walter Anthony 1, Thomas Schneider von Deimling2,3, Ingmar Nitze 3,4, Steve Frolking5, Abraham Emond6, Ronald Daanen6, Peter Anthony1, Prajna Lindgren7, Benjamin Jones1 & Guido Grosse 3,8 Permafrost carbon feedback (PCF) modeling has focused on gradual thaw of near-surface permafrost leading to enhanced carbon dioxide and methane emissions that accelerate global 1234567890():,; climate warming. These state-of-the-art land models have yet to incorporate deeper, abrupt thaw in the PCF. Here we use model data, supported by field observations, radiocarbon dating, and remote sensing, to show that methane and carbon dioxide emissions from abrupt thaw beneath thermokarst lakes will more than double radiative forcing from cir- cumpolar permafrost-soil carbon fluxes this century. Abrupt thaw lake emissions are similar under moderate and high representative concentration pathways (RCP4.5 and RCP8.5), but their relative contribution to the PCF is much larger under the moderate warming scenario. Abrupt thaw accelerates mobilization of deeply frozen, ancient carbon, increasing 14C- depleted permafrost soil carbon emissions by ~125–190% compared to gradual thaw alone. These findings demonstrate the need to incorporate abrupt thaw processes in earth system models for more comprehensive projection of the PCF this century. 1 Water and Environmental Research Center, University of Alaska Fairbanks, Fairbanks, AK 99775, USA. 2 Max Planck Institute for Meteorology, 20146 Hamburg, Germany. 3 Alfred Wegener Institute Helmholtz Centre for Polar and Marine Research, 14473 Potsdam, Germany. 4 Institute of Geography, University of Potsdam, 14476 Potsdam, Germany. -

Ecosystem Carbon Response of an Arctic Peatland to Simulated Permafrost Thaw
This is a self-archived version of an original article. This version may differ from the original in pagination and typographic details. Author(s): Voigt, Carolina; Marushchak, Maija; Mastepanov, Mikhail; Lamprecht, Richard E.; Christensen, Torben R.; Dorodnikov, Maxim; Jackowicz‐Korczyński, Marcin; Lindgren, Amelie; Lohila, Annalea; Nykänen, Hannu; Oinonen, Markku; Oksanen, Timo; Palonen, Vesa; Treat, Claire C.; Martikainen, Pertti J.; Biasi, Christina Title: Ecosystem carbon response of an Arctic peatland to simulated permafrost thaw Year: 2019 Version: Accepted version (Final draft) Copyright: © 2019 John Wiley & Sons Ltd. Rights: In Copyright Rights url: http://rightsstatements.org/page/InC/1.0/?language=en Please cite the original version: Voigt, C., Marushchak, M., Mastepanov, M., Lamprecht, R. E., Christensen, T. R., Dorodnikov, M., . Biasi, C. (2019). Ecosystem carbon response of an Arctic peatland to simulated permafrost thaw. Global Change Biology, 25 (5), 1746-1764. doi:10.1111/gcb.14574 DR. CAROLINA VOIGT (Orcid ID : 0000-0001-8589-1428) MS. CLAIRE C TREAT (Orcid ID : 0000-0002-1225-8178) Article type : Primary Research Articles Ecosystem carbon response of an Arctic peatland to simulated permafrost thaw Running head: Carbon fluxes of thawing permafrost peatlands 1,2* 2,3 4,5 2 Article Carolina Voigt , Maija E. Marushchak , Mikhail Mastepanov , Richard E. Lamprecht , Torben R. 4,5 6 4,5 5,7 Christensen , Maxim Dorodnikov , Marcin Jackowicz-Korczyński , Amelie Lindgren , Annalea Lohila8, Hannu Nykänen2, Markku Oinonen9, Timo -

Oxidation of Organic Material by Heterotrophic Bacteria Is An
To appear in “Processes in Microbial Ecology” To be published by Oxford University Press David L. Kirchman Draft: Feb 24, 2010 Chapter 5 Degradation of organic material The previous chapter discussed the synthesis of organic material by autotrophic microbes, the primary producers. This chapter will discuss the degradation of that organic material by heterotrophic microbes. These two processes are large parts of the natural carbon cycle. Nearly all of the 120 gigatons of carbon dioxide fixed each year into organic material by primary producers is returned back to the atmosphere by heterotrophic microbes, macroscopic animals and even autotrophic organisms (Chapter 4). Note the “nearly” in the last sentence. While primary production is mostly balanced by degradation, imbalances occur, affecting many aspects of the ecosystem. It is these imbalances that set whether the biota is a net producer or consumer of atmospheric carbon dioxide. Since these imbalances depend on degradation as well as primary production, so too do both primary production and organic matter degradation determine the net contribution of the biota to fluxes of carbon dioxide to and from the atmosphere. As with all biogeochemical cycles, the carbon cycle consists of reservoirs (concentrations or amounts of material) connected by fluxes (time-dependent rates) made of both natural and anthropogenic processes (Fig. 1). The natural rates of exchange between carbon reservoirs are much larger than the anthropogenetic ones. In particular, the natural production of carbon dioxide by heterotrophs is much higher than the anthropogenic production due to the burning of fossil fuels and other human activities. The problem is that, because the anthropogenetic production of carbon dioxide is not balanced by carbon dioxide consumption, concentrations in the atmosphere are increasing and our planet is warming up (Chapter 1). -

Arctic Climate Feedbacks: Global Implications
for a living planet ARCTIC CLIMATE FEEDBACKS: GLOBAL IMPLICATIONS ARCTIC CLIMATE FEEDBACKS: GLOBAL IMPLICATIONS Martin Sommerkorn & Susan Joy Hassol, editors With contributions from: Mark C. Serreze & Julienne Stroeve Cecilie Mauritzen Anny Cazenave & Eric Rignot Nicholas R. Bates Josep G. Canadell & Michael R. Raupach Natalia Shakhova & Igor Semiletov CONTENTS Executive Summary 5 Overview 6 Arctic Climate Change 8 Key Findings of this Assessment 11 1. Atmospheric Circulation Feedbacks 17 2. Ocean Circulation Feedbacks 28 3. Ice Sheets and Sea-level Rise Feedbacks 39 4. Marine Carbon Cycle Feedbacks 54 5. Land Carbon Cycle Feedbacks 69 6. Methane Hydrate Feedbacks 81 Author Team 93 EXECUTIVE SUMMARY VER THE PAST FEW DECADES, the Arctic has warmed at about twice the rate of the rest of the globe. Human-induced climate change has Oaffected the Arctic earlier than expected. As a result, climate change is already destabilising important arctic systems including sea ice, the Greenland Ice Sheet, mountain glaciers, and aspects of the arctic carbon cycle including altering patterns of frozen soils and vegetation and increasing “Human-induced methane release from soils, lakes, and climate change has wetlands. The impact of these changes on the affected the Arctic Arctic’s physical systems, earlier than expected.” “There is emerging evidence biological and growing concern that systems, and human inhabitants is large and projected to grow arctic climate feedbacks throughout this century and beyond. affecting the global climate In addition to the regional consequences of arctic system are beginning climate change are its global impacts. Acting as the to accelerate warming Northern Hemisphere’s refrigerator, a frozen Arctic plays a central role in regulating Earth’s climate signifi cantly beyond system. -

The Microbial Carbon Pump Concept: Potential Biogeochemical Significance in the Globally Changing Ocean Louis Legendre, Richard B
The microbial carbon pump concept: Potential biogeochemical significance in the globally changing ocean Louis Legendre, Richard B. Rivkin, Markus Weinbauer, Lionel Guidi, Julia Uitz To cite this version: Louis Legendre, Richard B. Rivkin, Markus Weinbauer, Lionel Guidi, Julia Uitz. The microbial carbon pump concept: Potential biogeochemical significance in the globally changing ocean. Progress in Oceanography, Elsevier, 2015, 134, pp.432-450. 10.1016/j.pocean.2015.01.008. hal-01120262 HAL Id: hal-01120262 https://hal.sorbonne-universite.fr/hal-01120262 Submitted on 25 Feb 2015 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Review The microbial carbon pump concept: potential biogeochemical significance in the globally changing ocean POSSIBLE ALTERNATIVE TITLE The microbial carbon pump concept: potential biogeochemical significance in the context of the biological and carbonate pumps and in the globally changing ocean Louis Legendrea,b,*, Richard B. Rivkinc, Markus Weinbauera,b, Lionel Guidia,b, Julia Uitza,b a Sorbonne Universités, UPMC Univ. Paris 06, UMR 7093, Laboratoire d'Océanographie de Villefranche, 06230 Villefranche-sur-Mer, France b CNRS, UMR 7093, Laboratoire d'Océanographie de Villefranche, 06230 Villefranche-sur- Mer, France c Department of Ocean Sciences, Memorial University of Newfoundland, St. -

Analytically Tractable Climate-Carbon Cycle Feedbacks Under 21St Century Anthropogenic Forcing Steven J
Earth Syst. Dynam. Discuss., https://doi.org/10.5194/esd-2017-78 Manuscript under review for journal Earth Syst. Dynam. Discussion started: 12 September 2017 c Author(s) 2017. CC BY 4.0 License. Analytically tractable climate-carbon cycle feedbacks under 21st century anthropogenic forcing Steven J. Lade1,2,3, Jonathan F. Donges1,4, Ingo Fetzer1,3, John M. Anderies5, Christian Beer3,6, Sarah E. Cornell1, Thomas Gasser7, Jon Norberg1, Katherine Richardson8, Johan Rockström1, and Will Steffen1,2 1Stockholm Resilience Centre, Stockholm University, Stockholm, Sweden 2Fenner School of Environment and Society, The Australian National University, Canberra, Australian Capital Territory, Australia 3Bolin Centre for Climate Research, Stockholm University, Stockholm, Sweden 4Potsdam Institute for Climate Impact Research, Potsdam, Germany 5School of Sustainability and School of Human Evolution and Social Change, Arizona State University, Tempe, Arizona, USA 6Department of Environmental Science and Analytical Chemistry (ACES), Stockholm University, Stockholm, Sweden 7International Institute for Applied Systems Analysis, Laxenburg, Austria 8Center for Macroecology, Evolution, and Climate, University of Copenhagen, Natural History Museum of Denmark, Copenhagen, Denmark Correspondence to: Steven Lade ([email protected]) Abstract. Changes to climate-carbon cycle feedbacks may significantly affect the Earth System’s response to greenhouse gas emissions. These feedbacks are usually analysed from numerical output of complex and arguably opaque Earth System Models (ESMs). Here, we construct a stylized global climate-carbon cycle model, test its output against complex ESMs, and investigate the strengths of its climate-carbon cycle feedbacks analytically. The analytical expressions we obtain aid understanding of 5 carbon-cycle feedbacks and the operation of the carbon cycle. -
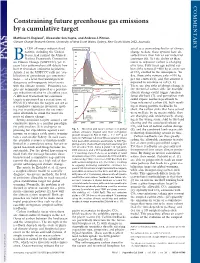
Constraining Future Greenhouse Gas Emissions by a Cumulative Target
COMMENTARY Constraining future greenhouse gas emissions by a cumulative target Matthew H. England1, Alexander Sen Gupta, and Andrew J. Pitman Climate Change Research Centre, University of New South Wales, Sydney, New South Wales 2052, Australia y 1994 all major industrialized Emissions (PgC yr-1) acted as a tremendous buffer of climate nations, including the United 16 6 change to date; these systems have ab- 1 States, had ratified the United 14 sorbed more than half of our industrial emissions (6). Yet the ability of these Nations Framework Convention 12 Bon Climate Change (UNFCCC), yet 15 2 stores to sequester carbon is changing years later policymakers still debate how 10 over time. Fifty years ago natural car- best to formulate emissions legislation. 8 bon sinks removed Ϸ600 kg of every ton Article 2 of the UNFCCC calls for ‘‘sta- 6 of CO2 emitted to the atmosphere. To- bilization of greenhouse gas concentra- 4 day, these sinks remove only Ϸ550 kg 4 5 per ton emitted (6), and this amount is tions...atalevel that would prevent 2 3 dangerous anthropogenic interference 950 PgC expected to continue to fall (3, 4). with the climate system.’’ Emissions tar- 1950 2000 2050 2100 2150 There are also risks of abrupt change in Cummulative Emissions gets are commonly quoted as a percent- 6 1785 PgC the terrestrial carbon sink; for example, relative to 2000 (PgC) in 2200 5 700 PgC age reduction relative to a baseline year. 590 PgC climate change could trigger Amazon 600 1 3 66% probability forest die-back (7), and permafrost melt A different framework for emissions 2 of not exceeding 2o warming targets is presented in a recent issue of 400 590 PgC could expose northern peatlands to PNAS (1), wherein the targets are set as large releases of carbon (8), both result- 200 170 PgC ing in strong positive feedbacks. -

Global Significance of the Changing Freshwater Carbon Cycle - Eos
9/22/2017 Global Significance of the Changing Freshwater Carbon Cycle - Eos Global Significance of the Changing Freshwater Carbon Cycle Freshwater ecosystems constitute a small fraction of our planet but play a disproportionately large and critical role in the global carbon cycle. Small inland freshwater bodies play a large role in the global carbon cycle. Here Ruddiman Lagoon, a small freshwater lake, joins Muskegon Lake, a freshwater estuary in Michigan that drains the extensive Muskegon River watershed into Lake Michigan. This North American Great Lake feeds the Saint Lawrence River, which drains into the Atlantic Ocean. Credit: Janet H. Vail By Bopaiah A. Biddanda 21 March 2017 As human activities continue to pump carbon into the atmosphere, the backbone of our understanding of the resulting warming is our knowledge of where that carbon is going: into the atmosphere, into the land, and into bodies of water. When it comes to accounting for the carbon absorbed and emitted by water, the role of inland freshwater may appear quite small compared to the vastness of Earth’s oceans. After all, inland lakes, rivers, streams, reservoirs, wetlands, and estuaries cover less than 4% of Earth’s surface [Downing, 2010; Verpoorter et al., 2014]. But recent research shows that the roughly 200 million bodies of inland water play a much larger role in the global carbon cycle than their small footprint suggests. Inland streams and rivers move vast amounts of carbon from the land to the ocean, acting as carbon’s busy transit system. They also play a disproportionately large role in the global carbon cycle through their high rates of carbon respiration and sequestration [Cole et al., 2007; Tranvik et al., 2009]. -

Cryogenic Soil Processes in a Changing Climate
Cryogenic soil processes in a changing climate Marina Becher Department of Ecology and Environmental Science Umeå, 2015 This work is protected by the Swedish Copyright Legislation (Act 1960:729) © Marina Becher 2015 ISBN: 978-91-7601-361-8 Cover: Non-sorted circles at Mount Suorooaivi, photo by Marina Becher Printed by: KBC Service center Umeå, Sweden 2015 ”Conclusions regarding origin are that: (1) the origin of most forms of patterned ground is uncertain: (2)…” From Abstract in Washburn, A. L., 1956. "Classification of patterned ground and review of suggested origins." Geological Society of America Bulletin 67(7): 823-866. Table of Contents Table of Contents i List of papers iii Author contributions iv Abstract v Introduction 1 Background 1 Cryoturbation and its impact on the carbon balance 1 Cryogenic disturbances and their effect on plants 2 Cryoturbation within non-sorted circles 2 Objective of the thesis 4 Study site 4 Methods 6 Results and Discussion 7 Over what timescale does cryoturbation occur in non-sorted circles? 7 What effect does cryogenic disturbance have on vegetation? 8 What is the effect of cryogenic disturbances on carbon fluxes? 10 Conclusion 11 Acknowledgements 11 References 12 TACK! 17 i ii List of papers This thesis is a summary of the following four studies, which will be referred to by their roman numerals. I. Becher, M., C. Olid and J. Klaminder 2012. Buried soil organic inclusions in non-sorted circles fields in northern Sweden: Age and Paleoclimatic context. Journal of geophysical research: Biogeosciences 118:104-111 II. Becher, M., N. Börlin and J. Klaminder.