The Malacca Strait Separates Distinct Faunas of Poorly-Flying Cautires Net
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Thomas Tew and Pirate Settlements of the Indo - Atlantic Trade World, 1645 -1730 1 Kevin Mcdonald Department of History University of California, Santa Cruz
‘A Man of Courage and Activity’: Thomas Tew and Pirate Settlements of the Indo - Atlantic Trade World, 1645 -1730 1 Kevin McDonald Department of History University of California, Santa Cruz “The sea is everything it is said to be: it provides unity, transport , the means of exchange and intercourse, if man is prepared to make an effort and pay a price.” – Fernand Braudel In the summer of 1694, Thomas Tew, an infamous Anglo -American pirate, was observed riding comfortably in the open coach of New York’s only six -horse carriage with Benjamin Fletcher, the colonel -governor of the colony. 2 Throughout the far -flung English empire, especially during the seventeenth century, associations between colonial administrators and pirates were de rig ueur, and in this regard , New York was similar to many of her sister colonies. In the developing Atlantic world, pirates were often commissioned as privateers and functioned both as a first line of defense against seaborne attack from imperial foes and as essential economic contributors in the oft -depressed colonies. In the latter half of the seventeenth century, moreover, colonial pirates and privateers became important transcultural brokers in the Indian Ocean region, spanning the globe to form an Indo-Atlantic trade network be tween North America and Madagascar. More than mere “pirates,” as they have traditionally been designated, these were early modern transcultural frontiersmen: in the process of shifting their theater of operations from the Caribbean to the rich trading grounds of the Indian Ocean world, 1 An earlier version of this paper was presented at the “Counter -Currents and Mainstreams in World History” conference at UCLA on December 6-7, 2003, organized by Richard von Glahn for the World History Workshop, a University of California Multi -Campus Research Unit. -

Curculio a Newsletter Devoted to Dissemination of Knowledge About Curculionoidea
CURCULIO A NEWSLETTER DEVOTED TO DISSEMINATION OF KNOWLEDGE ABOUT CURCULIONOIDEA NO. 41 - MARCH 1997 CANADIAN MUSEUM OF NATURE P.O. BOX 3443, STATION D EDITED BY OTTAWA, ON. K1P 6P4 ROBERT S. ANDERSON CANADA Mike Morris (retired) September 1992, Poland CURCULIO EDITORIAL COMMENTS Yes, this number of CURCULIO is late. It should have been prepared and sent out in September of 1996, but our museum has been going through a move to new facilities and things have been in turmoil. We are now settled in and most services are up and running again. The collections, which were packed away for the move, are also now accessible again. Seems that things went quite well and in fact our Entomology section fared quite well as the move into our new facilities resulted in a great deal more space than we already had. We’re now getting the laboratories prepared and should be back to full speed in the near future. Our mailing address has not changed! On another note, congratulations are in order for Rolf Oberprieler who has accepted the weevil systematics position at CSIRO in Canberra, Australia. This will be a great new challenge for Rolf and one in which we can all be sure he will fare admirably. The Australian weevil fauna is exceptionally diverse and interesting and with the recent books by Elwood Zimmerman, a great base has been set for further and more detailed work on the fauna. I understand Rolf will be there sometime this summer. Now all he has to do is master ‘G’Day’ and ‘mate’ and he’ll have it made! Best wishes to Rolf and his family. -

The 20 February 2010 Madeira Flash-Floods
Nat. Hazards Earth Syst. Sci., 12, 715–730, 2012 www.nat-hazards-earth-syst-sci.net/12/715/2012/ Natural Hazards doi:10.5194/nhess-12-715-2012 and Earth © Author(s) 2012. CC Attribution 3.0 License. System Sciences The 20 February 2010 Madeira flash-floods: synoptic analysis and extreme rainfall assessment M. Fragoso1, R. M. Trigo2, J. G. Pinto3, S. Lopes1,4, A. Lopes1, S. Ulbrich3, and C. Magro4 1IGOT, University of Lisbon, Portugal 2IDL, Faculty of Sciences, University of Lisbon, Portugal 3Institute for Geophysics and Meteorology, University of Cologne, Germany 4Laboratorio´ Regional de Engenharia Civil, R.A. Madeira, Portugal Correspondence to: M. Fragoso ([email protected]) Received: 9 May 2011 – Revised: 26 September 2011 – Accepted: 31 January 2012 – Published: 23 March 2012 Abstract. This study aims to characterise the rainfall ex- island of Madeira is quite densely populated, particularly in ceptionality and the meteorological context of the 20 Febru- its southern coast, with circa 267 000 inhabitants in 2011 ary 2010 flash-floods in Madeira (Portugal). Daily and census, with 150 000 (approximately 40 %) living in the Fun- hourly precipitation records from the available rain-gauge chal district, one of the earliest tourism hot spots in Europe, station networks are evaluated in order to reconstitute the currently with approximately 30 000 hotel beds. In 2009, the temporal evolution of the rainstorm, as its geographic inci- archipelago received 1 million guests; this corresponds to an dence, contributing to understand the flash-flood dynamics income of more than 255 Million Euro (INE-Instituto Na- and the type and spatial distribution of the associated im- cional de Estat´ıstica, http://www.ine.pt). -

The Phylogenetic Structure of Metriorrhynchini Fauna of Sulawesi
Zoological Studies 50(5): 645-656 (2011) The Phylogenetic Structure of Metriorrhynchini Fauna of Sulawesi (Coleoptera: Lycidae) with Descriptions of a New Genus, Mangkutanus, and Three New Species of Xylobanus Vaclav Kubecek, Milan Dvorak, and Ladislav Bocak* Department of Zoology, Faculty of Science, Palacky University, Tr. Svobody 26, 771 46 Olomouc, Czech Republic (Accepted May 20, 2011) Vaclav Kubecek, Milan Dvorak, and Ladislav Bocak (2011) The phylogenetic structure of Metriorrhynchini fauna of Sulawesi (Coleoptera: Lycidae) with descriptions of a new genus, Mangkutanus, and three new species of Xylobanus. Zoological Studies 50(5): 645-656. The phylogenetic structure of the Metriorrhynchini fauna from Sulawesi was investigated. We obtained DNA sequences for 3 fragments of the mitochondrial genome: cytochrome oxidase I, tRNA-Leu, and cytochrome oxidase subunit 2; NADH dehydrogenase subunit 5, tRNA- Ser, tRNA-Glu, and tRNA-Phe; and rrnl, tRNA-Leu, and NADH dehydrogenase subunit 1 for a total of 3070 aligned nucleotide positions, and performed a phylogenetic analysis of 12 genera of the Metriorrhynchini from Sulawesi and adjacent regions. Metriorrhynchini formed a monophylum that was split into 2 clades: an Afro- Oriental clade consisting of Cautires Waterhouse, 1879 and Xylobanus Waterhouse, 1879, and an Austral- Oriental clade formed by 10 genera distributed either only in the Australian Region, or with hundreds of species in Australia and New Guinea and a few in the Oriental region. Altogether, 11 genera are known from Sulawesi, four of them endemic to the island, only 2 genera, Xylobanus and Cautires, with a presumed Oriental origin, and 9 belonging to the Australian clade. All 87 metriorrhynchine species except 1 are endemic to Sulawesi, and 79 (91%) of them belong to the Australian lineage. -
Austromonticola, a New Genus of Broad-Nosed Weevil (Coleoptera, Curculionidae, Entiminae) from Montane Areas of New Zealand
A peer-reviewed open-access journal ZooKeys 707: 73–130 (2017) Revision of Austromonticola weevils 73 doi: 10.3897/zookeys.707.12649 MONOGRAPH http://zookeys.pensoft.net Launched to accelerate biodiversity research Austromonticola, a new genus of broad-nosed weevil (Coleoptera, Curculionidae, Entiminae) from montane areas of New Zealand Samuel D. J. Brown1,2 1 Bio-Protection Research Centre, PO Box 85084, Lincoln University 7647, Canterbury, New Zealand 2 AgResearch, Gerald St, Lincoln, Canterbury, New Zealand Corresponding author: Samuel D. J. Brown ([email protected]) Academic editor: M. Alonso-Zarazaga | Received 17 March 2017 | Accepted 20 August 2017 | Published 10 October 2017 http://zoobank.org/0DF0C91D-3B1D-450D-80F3-F32F8EE7801D Citation: Brown SDJ (2017) Austromonticola, a new genus of broad-nosed weevil (Coleoptera, Curculionidae, Entiminae) from montane areas of New Zealand. ZooKeys 707: 73–130. https://doi.org/10.3897/zookeys.707.12649 Abstract Austromonticola gen. n. is proposed for a group of eight New Zealand alpine broad-nosed weevil species, all of which are here described: A. atriarius sp. n. (type locality: Umbrella Mountains, Central Otago), A. caelibatus sp. n. (type locality: Ohau Range, Mackenzie), A. furcatus sp. n. (type locality: Old Man Range, Central Otago), A. inflatus sp. n. (type locality: Hawkdun Range, Central Otago), A. planulatus sp. n. (type locality: St Marys Range, Central Otago), A. postinventus sp. n. (type locality: Kirkliston Range, South Canterbury), A. mataura sp. n. (type locality: Mt Dick, Otago Lakes) and A. rotundus sp. n. (type locality: Old Man Range, Central Otago). All species occur exclusively above 1000 m elevation in the mountains of Central Otago and South Canterbury in the South Island. -

Conspicuousness, Phylogenetic Structure, and Origins of Müllerian
www.nature.com/scientificreports OPEN Conspicuousness, phylogenetic structure, and origins of Müllerian mimicry in 4000 lycid beetles from all zoogeographic regions Michal Motyka1, Dominik Kusy1, Michal Masek1, Matej Bocek1, Yun Li1, R. Bilkova1, Josef Kapitán2, Takashi Yagi3 & Ladislav Bocak1* Biologists have reported on the chemical defences and the phenetic similarity of net-winged beetles (Coleoptera: Lycidae) and their co-mimics. Nevertheless, our knowledge has remained fragmental, and the evolution of mimetic patterns has not been studied in the phylogenetic context. We illustrate the general appearance of ~ 600 lycid species and ~ 200 co-mimics and their distribution. Further, we assemble the phylogeny using the transcriptomic backbone and ~ 570 species. Using phylogenetic information, we closely scrutinise the relationships among aposematically coloured species, the worldwide diversity, and the distribution of aposematic patterns. The emitted visual signals difer in conspicuousness. The uniform coloured dorsum is ancestral and was followed by the evolution of bicoloured forms. The mottled patterns, i.e. fasciate, striate, punctate, and reticulate, originated later in the course of evolution. The highest number of sympatrically occurring patterns was recovered in New Guinea and the Andean mountain ecosystems (the areas of the highest abundance), and in continental South East Asia (an area of moderate abundance but high in phylogenetic diversity). Consequently, a large number of co-existing aposematic patterns in a single region and/or locality is the rule, in contrast with the theoretical prediction, and predators do not face a simple model-like choice but cope with complex mimetic communities. Lycids display an ancestral aposematic signal even though they sympatrically occur with diferently coloured unproftable relatives. -

Analysis of Y-Chromosome and Mtdna Variability in the Madeira Archipelago Population
International Congress Series 1288 (2006) 94–96 www.ics-elsevier.com Analysis of Y-chromosome and mtDNA variability in the Madeira Archipelago population Ana T. Fernandes *, Rita Gonc¸alves, Alexandra Rosa, Anto´nio Brehm Human Genetics Laboratory, University of Madeira, Campus of Penteada, Funchal, Portugal Abstract. The Atlantic archipelago of Madeira is made up of two islands (Madeira and Porto Santo) with 250,000 inhabitants. These islands were discovered and settled by the Portuguese in the 15th century and played an important role in the complex Atlantic trade network in the following centuries. The genetic composition of the Madeira Islands’ population was investigated by analyzing Y-chromosomal bi-allelic and STR markers in three different regions of the main island plus Porto Santo. We compared the results with mtDNA data and used the Y-chromosome STRs to determine the variability within each haplogroup. A sample of 142 unrelated males divided into four groups (Funchal City, West Madeira, North and East Madeira and Porto Santo) were analyzed. Significant genetic differences between these regions and the population of Funchal were found. The population of Funchal had lower gene diversity than expected. D 2006 Elsevier B.V. All rights reserved. Keywords: Madeira Island; Y-chromosome; mtDNA 1. Introduction The Madeira archipelago is made up of two inhabited islands, Madeira and Porto Santo, and has a population of about 250,000 inhabitants, with more than half living in Funchal. The Portuguese colonized the Madeira Archipelago in the 15th century and, in the beginning of the colonization, the archipelago was divided into three parts (Southwest and Northeast in the Madeira Island, and Porto Santo) and given to three administrators [1]. -

Chapter I the Portuguese Empire
Decay or defeat ? : an inquiry into the Portuguese decline in Asia 1580-1645 Veen, Ernst van Citation Veen, E. van. (2000, December 6). Decay or defeat ? : an inquiry into the Portuguese decline in Asia 1580-1645. Research School of Asian, African, and Amerindian Studies (CNWS), Leiden University. Retrieved from https://hdl.handle.net/1887/15783 Version: Not Applicable (or Unknown) License: Downloaded from: https://hdl.handle.net/1887/15783 Note: To cite this publication please use the final published version (if applicable). CHAPTER I THE PORTUGUESE EMPIRE The boundaries Until well into the seventeenth century, as far as the Iberians were concerned, the way the world was divided and the role they were to play therein as champions of the church was clear-cut and straightforward. Already in the fifteenth century the rights of the Portuguese monarchs on the portus, insulas, terras et maria still to be conquered had been confirmed by Papal edicts. They bestowed the privilege to intrude into the countries of the Saracenes and heathens, to take them prisoner, take all their possessions and reduce them to eternal slavery. Derived from this right of conquest were the rights of legislation, jurisdiction and tribute and the monopolies of navigation, trade and fishing. Besides, the kings were allowed to build churches, cloisters and other holy places and to send clergy and other volunteers, to spread the true religion, to receive confessions and to give absolutions. Excommunication or interdiction were the penalties for Christians who violated these royal monopolies.1 As the Castilians were just as keen on the collection of slaves and gold and the overseas expansion of the mission, a clash of interests was inevitable.2 In 1479, the Castilians used the opportunity of king Afonso V's defeat, after he attempted to acquire the Castilian throne, to establish their rights on the Canary islands. -

Madeira and the Beginnings of New World Sugar Cane Cultivation and Plantation Slavery: a Study in Institution Building*
MADEIRA AND THE BEGINNINGS OF NEW WORLD SUGAR CANE CULTIVATION AND PLANTATION SLAVERY: A STUDY IN INSTITUTION BUILDING* Sidney M. Greenfield Department of Anthropology The University of Wisconsin-Milwaukee Milwaukee, Wisconsin 53201 Although the tropical New World is generally taken as the context for the comparative and historical study of plantation systems and of slavery, the institutional complex of the slave plantation' did not have its origins in the Western Hemisphere. Instead, as Charles Verlinden2 and others have demon- strated so ably, the culture complex can be traced back to the European colonies established in Palestine following the First Crusade. There sugar was grown by a mixed force of free and slave workers. The sugar then was exported, with some of it reaching the developing markets of Western Europe. Sugar cane had been known to Europe at least as far back as antiquity, although its distribution was limited. The cultivation of the crop, and its in- creased availability and wider distribution, came to Mediterranean Europe in the wake of the expansion of Islam. Sugar cane, most probably first domesticated in India, had been grown in Asia Minor and was carried along with the spread of Islam to the coastal regions of North Africa, the islands of the Mediterranean, and eventually to the Moorish kingdoms in the Iberian peninsula.3 Given the intensity of the conflict between Christianity and Islam during the Middle Ages, trade between the areas controlled by the rival religions tended to be minimal. Hence sugars produced in areas under Moslem control went, for the most part, to supply markets in the Near East and North Africa, while the markets of Europe had to be supplied from areas under Christian control. -
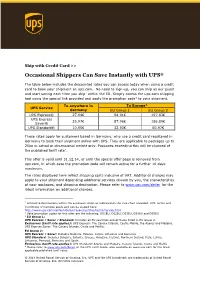
Occasional Shippers Can Save Instantly with UPS®
Ship with Credit Card >> Occasional Shippers Can Save Instantly with UPS® The table below includes the discounted rates you can access today when using a credit card to book your shipment on ups.com. No need to sign-up, you can ship as our guest and start saving each time you ship1 within the EU. Simply access the ups.com shipping tool using the special link provided and apply the promotion code2 to your shipment. To anywhere in To Europe3 UPS Service Germany EU Group 1 EU Group 2 UPS Express® 27.94€ 94.01€ 197.83€ UPS Express 25.97€ 87.96€ 186.89€ Saver® UPS Standard® 10.95€ 32.93€ 50.97€ These rates apply for customers based in Germany, who use a credit card registered in Germany to book their shipment online with UPS. They are applicable to packages up to 20kg in actual or dimensional weight only. Packages exceeding this will be charged at the published tariff rate4. This offer is valid until 31.12.14, or until the special offer page is removed from ups.com, in which case the promotion code will remain active for a further 10 days maximum. The rates displayed here reflect shipping costs inclusive of VAT. Additional charges may apply to your shipment depending additional services chosen by you, the characteristics of your packages, and shipping destination. Please refer to www.ups.com/de/en for the latest information on additional charges. 1 Limited to destinations within the European Union as indicated on the rate chart provided. UPS Terms and Conditions of Carriage apply and can be viewed here: http://www.ups.com/content/de/en/resources/ship/terms/service.html 2 Valid promotion codes for this offer are the following: DE1EU, DE2EU, DE3EU, DE4EU and DE5EU 3 EU Group 1: UPS Express / Saver / Standard: Includes all EU countries except those listed in EU Group 2. -

Socotra Archipelago – a Lifeboat in the Sea of Changes: Advancement in Socotran Insect Biodiversity Survey
ACTA ENTOMOLOGICA MUSEI NATIONALIS PRAGAE Published 17.xii.2012 Volume 52 (supplementum 2), pp. 1–26 ISSN 0374-1036 Socotra Archipelago – a lifeboat in the sea of changes: advancement in Socotran insect biodiversity survey Jan BATELKA Nad Vodovodem 16, CZ-100 00 Prague 10, Czech Republic; e-mail: [email protected] Abstract. Nature conditions in the Socotra Archipelago are briefl y summarised, main factors contributing to the composition and diversity of the insect fauna, i.e. geological history, geographic position, geomorphology, climatic conditions and plant diversity, are circumscribed. Results of the Socotran invertebrate biodiversity research are reviewed, the fi rst annotated list of 40 insect genera and subgenera endemic to the Socotra Archipelago is provided. Recorded high level of ende- mism in the Socotra Island is in accordance with the estimated geological age and continuous stability of its ecosystem. Brief comparison of biodiversity in Socotra and Seychelles Islands – both granitic archipelagos of Gondwanan origin in the West Indian Ocean – is performed. Difference in composition of insect fauna in Socotra and allied Abd el Kuri Island is commented. Main threats and conservation issues concerning the fragile Socotran ecosystem are summarised, and examples of possible consequences for insect fauna are given. Based on comparison of scarce outputs and research activities concerning the Socotran insect fauna so far with the number of those elaborated for other countries/islands, establishing of a long-term insect survey is recommended. Key words. Review, insularity, geology, fl ora, climate, biodiversity, endemism, insect genera, habitat conservation, insect survey, Yemen, Socotra Introduction Because of their remarkable richness in endemic forms, study of the isolated biotas (also called ‘insularity’) has been one of the most interesting topics among naturalists and especially evolutionary biologists since the second half of the 19th century (WITT & MALIAKAL-WITT 2007). -

Molecular Phylogenetics of the Superfamily Curculionoidea (Insecta: Coleoptera)
Molecular Phylogenetics of the Superfamily Curculionoidea (Insecta: Coleoptera) Conrad Paulus Dias Trafford Gillett A thesis submitted in fulfilment of the requirements for the degree of Doctor of Philosophy University of East Anglia Norwich, Norfolk, England March 2014 © This copy of the thesis has been supplied on condition that anyone who consults it is understood to recognise that its copyright rests with the author and that use of any information derived there-from must be in accordance with current UK Copyright Law. In addition, any quotation or extract must include full attribution. 1 Molecular Phylogenetics of the Superfamily Curculionoidea (Insecta: Coleoptera) Conrad Paulus Dias Trafford Gillett March 2014 Thesis abstract This thesis examines higher-level evolutionary history within the superfamily Curculionoidea, the most speciose family-level taxon, which includes beetles commonly known as weevils. This is achieved using a phylogenetic approach incorporating the largest datamatrix yet employed for weevil molecular systematics, and includes an investigation into the prospect of obtaining short phylogenetically informative amplicons from archival museum specimens. Newly obtained DNA sequence data is analysed from a variety of mitochondrial and nuclear loci, including 92 mitogenomes assembled through the approach of next-generation sequencing of pooled genomic DNA. The resulting trees are used to test previous morphological- and molecular-based hypotheses of weevil relationships and classification schemes. Mitogenomic-derived trees reveal topologies that are highly congruent with previous molecular studies, but that conflict with some morphological hypotheses. Strong nodal support strengthens inferences into the relationships amongst most weevil families and suggests that the largest family, the Curculionidae, is monophyletic, if the subfamily Platypodinae is excluded.